+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12542 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human CTLH-WDR26 supramolecular assembly | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
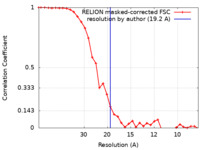
| Method | single particle reconstruction / cryo EM / Resolution: 19.2 Å | |||||||||
 Authors Authors | Chrustowicz J / Sherpa D / Schulman BA | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2021 Journal: Mol Cell / Year: 2021Title: GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme. Authors: Dawafuti Sherpa / Jakub Chrustowicz / Shuai Qiao / Christine R Langlois / Laura A Hehl / Karthik Varma Gottemukkala / Fynn M Hansen / Ozge Karayel / Susanne von Gronau / J Rajan Prabu / ...Authors: Dawafuti Sherpa / Jakub Chrustowicz / Shuai Qiao / Christine R Langlois / Laura A Hehl / Karthik Varma Gottemukkala / Fynn M Hansen / Ozge Karayel / Susanne von Gronau / J Rajan Prabu / Matthias Mann / Arno F Alpi / Brenda A Schulman /  Abstract: How are E3 ubiquitin ligases configured to match substrate quaternary structures? Here, by studying the yeast GID complex (mutation of which causes deficiency in glucose-induced degradation of ...How are E3 ubiquitin ligases configured to match substrate quaternary structures? Here, by studying the yeast GID complex (mutation of which causes deficiency in glucose-induced degradation of gluconeogenic enzymes), we discover supramolecular chelate assembly as an E3 ligase strategy for targeting an oligomeric substrate. Cryoelectron microscopy (cryo-EM) structures show that, to bind the tetrameric substrate fructose-1,6-bisphosphatase (Fbp1), two minimally functional GID E3s assemble into the 20-protein Chelator-GID, which resembles an organometallic supramolecular chelate. The Chelator-GID assembly avidly binds multiple Fbp1 degrons so that multiple Fbp1 protomers are simultaneously ubiquitylated at lysines near the allosteric and substrate binding sites. Importantly, key structural and biochemical features, including capacity for supramolecular assembly, are preserved in the human ortholog, the CTLH E3. Based on our integrative structural, biochemical, and cell biological data, we propose that higher-order E3 ligase assembly generally enables multipronged targeting, capable of simultaneously incapacitating multiple protomers and functionalities of oligomeric substrates. #1:  Journal: Biorxiv / Year: 2021 Journal: Biorxiv / Year: 2021Title: GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme Authors: Sherpa D / Chrustowicz J / Qiao S / Langlois CR / Hehl LA / Gottemukkala KV / Hansen FM / Karayel O / Prabu JR / Mann M / Alpi AF / Schulman BA | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12542.map.gz emd_12542.map.gz | 780.2 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12542-v30.xml emd-12542-v30.xml emd-12542.xml emd-12542.xml | 13.9 KB 13.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12542_fsc.xml emd_12542_fsc.xml | 3.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_12542.png emd_12542.png | 55.7 KB | ||
| Masks |  emd_12542_msk_1.map emd_12542_msk_1.map | 2.6 MB |  Mask map Mask map | |
| Others |  emd_12542_half_map_1.map.gz emd_12542_half_map_1.map.gz emd_12542_half_map_2.map.gz emd_12542_half_map_2.map.gz | 1.5 MB 1.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12542 http://ftp.pdbj.org/pub/emdb/structures/EMD-12542 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12542 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12542 | HTTPS FTP |
-Validation report
| Summary document |  emd_12542_validation.pdf.gz emd_12542_validation.pdf.gz | 335.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12542_full_validation.pdf.gz emd_12542_full_validation.pdf.gz | 334.3 KB | Display | |
| Data in XML |  emd_12542_validation.xml.gz emd_12542_validation.xml.gz | 9.3 KB | Display | |
| Data in CIF |  emd_12542_validation.cif.gz emd_12542_validation.cif.gz | 11.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12542 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12542 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12542 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12542 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12542.map.gz / Format: CCP4 / Size: 2.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12542.map.gz / Format: CCP4 / Size: 2.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.3571 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12542_msk_1.map emd_12542_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_12542_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_12542_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : CTLH-WDR26 complex comprising RANBP9-TWA1-ARMC8-RMND5A-MAEA-WDR26
| Entire | Name: CTLH-WDR26 complex comprising RANBP9-TWA1-ARMC8-RMND5A-MAEA-WDR26 |
|---|---|
| Components |
|
-Supramolecule #1: CTLH-WDR26 complex comprising RANBP9-TWA1-ARMC8-RMND5A-MAEA-WDR26
| Supramolecule | Name: CTLH-WDR26 complex comprising RANBP9-TWA1-ARMC8-RMND5A-MAEA-WDR26 type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Molecular weight | Theoretical: 1.3 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 75.36 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)