[English] 日本語
 Yorodumi
Yorodumi- EMDB-12003: CryoEM structure of the human sodium proton exchanger NHA2 in nanodisc -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12003 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of the human sodium proton exchanger NHA2 in nanodisc | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | secondary active transporter / membrane transport / ion antiport / pH regulation / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationlithium:proton antiporter activity / lithium ion transport / positive regulation of osteoclast development / sperm principal piece / sodium ion homeostasis / flagellated sperm motility / sodium:proton antiporter activity / regulation of insulin secretion involved in cellular response to glucose stimulus / clathrin-dependent endocytosis / inorganic cation transmembrane transport ...lithium:proton antiporter activity / lithium ion transport / positive regulation of osteoclast development / sperm principal piece / sodium ion homeostasis / flagellated sperm motility / sodium:proton antiporter activity / regulation of insulin secretion involved in cellular response to glucose stimulus / clathrin-dependent endocytosis / inorganic cation transmembrane transport / sodium ion transport / mitochondrial membrane / recycling endosome / Stimuli-sensing channels / synaptic vesicle membrane / recycling endosome membrane / monoatomic ion transmembrane transport / basolateral plasma membrane / mitochondrial inner membrane / apical plasma membrane / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
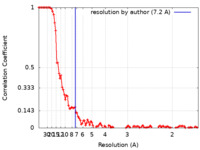
| Method | single particle reconstruction / cryo EM / Resolution: 7.2 Å | |||||||||
 Authors Authors | Woehlert D / Kuhlbrandt W | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: CryoEM structure of the human sodium proton exchanger NHA2 in nanodisc Authors: Woehlert D / Kuhlbrandt W / Yildiz O | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12003.map.gz emd_12003.map.gz | 100.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12003-v30.xml emd-12003-v30.xml emd-12003.xml emd-12003.xml | 13.4 KB 13.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12003_fsc.xml emd_12003_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_12003.png emd_12003.png | 44.7 KB | ||
| Masks |  emd_12003_msk_1.map emd_12003_msk_1.map | 107.2 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-12003.cif.gz emd-12003.cif.gz | 5.3 KB | ||
| Others |  emd_12003_half_map_1.map.gz emd_12003_half_map_1.map.gz emd_12003_half_map_2.map.gz emd_12003_half_map_2.map.gz | 99.2 MB 99.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12003 http://ftp.pdbj.org/pub/emdb/structures/EMD-12003 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12003 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12003 | HTTPS FTP |
-Validation report
| Summary document |  emd_12003_validation.pdf.gz emd_12003_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12003_full_validation.pdf.gz emd_12003_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_12003_validation.xml.gz emd_12003_validation.xml.gz | 18.5 KB | Display | |
| Data in CIF |  emd_12003_validation.cif.gz emd_12003_validation.cif.gz | 23.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12003 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12003 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12003 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12003 | HTTPS FTP |
-Related structure data
| Related structure data |  7b4mMC  7b4lC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12003.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12003.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.833 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12003_msk_1.map emd_12003_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_12003_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_12003_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human sodium proton exchanger NHA2 in nanodisc
| Entire | Name: Human sodium proton exchanger NHA2 in nanodisc |
|---|---|
| Components |
|
-Supramolecule #1: Human sodium proton exchanger NHA2 in nanodisc
| Supramolecule | Name: Human sodium proton exchanger NHA2 in nanodisc / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 150 KDa |
-Macromolecule #1: Sodium/hydrogen exchanger 9B2
| Macromolecule | Name: Sodium/hydrogen exchanger 9B2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 57.611672 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGDEDKRITY EDSEPSTGMN YTPSMHQEAQ EETVMKLKGI DANEPTEGSI LLKSSEKKLQ ETPTEANHVQ RLRQMLACPP HGLLDRVIT NVTIIVLLWA VVWSITGSEC LPGGNLFGII ILFYCAIIGG KLLGLIKLPT LPPLPSLLGM LLAGFLIRNI P VINDNVQI ...String: MGDEDKRITY EDSEPSTGMN YTPSMHQEAQ EETVMKLKGI DANEPTEGSI LLKSSEKKLQ ETPTEANHVQ RLRQMLACPP HGLLDRVIT NVTIIVLLWA VVWSITGSEC LPGGNLFGII ILFYCAIIGG KLLGLIKLPT LPPLPSLLGM LLAGFLIRNI P VINDNVQI KHKWSSSLRS IALSIILVRA GLGLDSKALK KLKGVCVRLS MGPCIVEACT SALLAHYLLG LPWQWGFILG FV LGAVSPA VVVPSMLLLQ GGGYGVEKGV PTLLMAAGSF DDILAITGFN TCLGIAFSTG STVFNVLRGV LEVVIGVATG SVL GFFIQY FPSRDQDKLV CKRTFLVLGL SVLAVFSSVH FGFPGSGGLC TLVMAFLAGM GWTSEKAEVE KIIAVAWDIF QPLL FGLIG AEVSIASLRP ETVGLCVATV GIAVLIRILT TFLMVCFAGF NLKEKIFISF AWLPKATVQA AIGSVALDTA RSHGE KQLE DYGMDVLTVA FLSILITAPI GSLLIGLLGP RLLQKVEHQN KDEEVQGETS VQV UniProtKB: Sodium/hydrogen exchanger 9B2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Number real images: 1251 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7b4m: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)