+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10404 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | FtsK motor domain bound to dsDNA, ADP-state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) (bacteria) / synthetic construct (others) / Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) (bacteria) / synthetic construct (others) /  Pseudomonas aeruginosa PAO1 (bacteria) / unidentified (others) Pseudomonas aeruginosa PAO1 (bacteria) / unidentified (others) | |||||||||
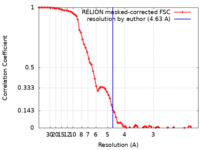
| Method | single particle reconstruction / cryo EM / Resolution: 4.63 Å | |||||||||
 Authors Authors | Jean NL / Lowe J | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2020 Journal: Proc Natl Acad Sci U S A / Year: 2020Title: FtsK in motion reveals its mechanism for double-stranded DNA translocation. Authors: Nicolas L Jean / Trevor J Rutherford / Jan Löwe /  Abstract: FtsK protein contains a fast DNA motor that is involved in bacterial chromosome dimer resolution. During cell division, FtsK translocates double-stranded DNA until both recombination sites are ...FtsK protein contains a fast DNA motor that is involved in bacterial chromosome dimer resolution. During cell division, FtsK translocates double-stranded DNA until both recombination sites are placed at mid cell for subsequent dimer resolution. Here, we solved the 3.6-Å resolution electron cryo-microscopy structure of the motor domain of FtsK while translocating on its DNA substrate. Each subunit of the homo-hexameric ring adopts a unique conformation and one of three nucleotide states. Two DNA-binding loops within four subunits form a pair of spiral staircases within the ring, interacting with the two DNA strands. This suggests that simultaneous conformational changes in all ATPase domains at each catalytic step generate movement through a mechanism related to filament treadmilling. While the ring is only rotating around the DNA slowly, it is instead the conformational states that rotate around the ring as the DNA substrate is pushed through. #1:  Journal: Biorxiv Journal: BiorxivTitle: FtsK in motion reveals its mechanism for double-stranded DNA translocation Authors: Jean NL / Rutherford TJ / Lowe J | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10404.map.gz emd_10404.map.gz | 4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10404-v30.xml emd-10404-v30.xml emd-10404.xml emd-10404.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10404_fsc.xml emd_10404_fsc.xml | 7.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_10404.png emd_10404.png | 150.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10404 http://ftp.pdbj.org/pub/emdb/structures/EMD-10404 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10404 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10404 | HTTPS FTP |
-Validation report
| Summary document |  emd_10404_validation.pdf.gz emd_10404_validation.pdf.gz | 251.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10404_full_validation.pdf.gz emd_10404_full_validation.pdf.gz | 250.4 KB | Display | |
| Data in XML |  emd_10404_validation.xml.gz emd_10404_validation.xml.gz | 10 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10404 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10404 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10404 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10404 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10404.map.gz / Format: CCP4 / Size: 35.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10404.map.gz / Format: CCP4 / Size: 35.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : FtsK motor domain bound to dsDNA, ADP-state
| Entire | Name: FtsK motor domain bound to dsDNA, ADP-state |
|---|---|
| Components |
|
-Supramolecule #1: FtsK motor domain bound to dsDNA, ADP-state
| Supramolecule | Name: FtsK motor domain bound to dsDNA, ADP-state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: FtsK motor domain
| Supramolecule | Name: FtsK motor domain / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) (bacteria) Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) (bacteria) |
| Recombinant expression | Organism:  |
-Supramolecule #3: DNA
| Supramolecule | Name: DNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Recombinant expression | Organism: synthetic construct (others) |
-Macromolecule #1: DNA translocase FtsK
| Macromolecule | Name: DNA translocase FtsK / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MVPDRREQSK AKERLLEREE ALAKHMSERE KRPPPKIDPP PSPKAPEPSK RVLKEKQAPL FVDTAVEGTL PPLSLLDPAE VKQKSYSPES LEAMSRLLEI KLKEFGVEVS VDSVHPGPVI TRFEIQPAAG VKVSRISNLA KDLARSLAVI SVRVVEVIPG KTTVGIEIPN ...String: MVPDRREQSK AKERLLEREE ALAKHMSERE KRPPPKIDPP PSPKAPEPSK RVLKEKQAPL FVDTAVEGTL PPLSLLDPAE VKQKSYSPES LEAMSRLLEI KLKEFGVEVS VDSVHPGPVI TRFEIQPAAG VKVSRISNLA KDLARSLAVI SVRVVEVIPG KTTVGIEIPN EDRQMVRFSE VLSSPEYDEH KSTVPLALGH DIGGRPIITD LAKMPHLLVA GTTGSGKSVG VNAMLLSILF KSTPSEARLI MIDPKMLELS IYEGIPHLLC PVVTDMKEAA NALRWSVAEM ERRYRLMAAM GVRNLAGFNR KVKDAEEAGT PLTDPLFRRE SPDDEPPQLS TLPTIVVVVD EFADMMMIVG KKVEELIARI AQKARAAGIH LILATQRPSV DVITGLIKAN IPTRIAFQVS SKIDSRTILD QGGAEQLLGH GDMLYLPPGT GLPIRVHGAF VSDDEVHRVV EAWKLRGAPD YIEDILAGVD EGGKLHHHHH H |
-Macromolecule #2: dsDNA substrate
| Macromolecule | Name: dsDNA substrate / type: dna / ID: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism: unidentified (others) |
| Sequence | String: ATATATATAT ATATATATAT |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Component:
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | ||||||||
| Details | 0.7 m/mL FtsK 1.5 uM 45 bp DNA |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 771 / Average electron dose: 39.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)