[English] 日本語
 Yorodumi
Yorodumi- EMDB-0342: CryoEM structure of Nav1.7 VSD2 (deactived state) in complex with... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0342 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Nav1.7 VSD2 (deactived state) in complex with the gating modifier toxin ProTx2 | |||||||||
 Map data Map data | Map generated with a mask including Nav (with truncated C-terminal coiled-coil), ProTx2 and Fv fragment. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | voltage-gated sodium channel / gating modifier toxin / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationdetection of mechanical stimulus involved in sensory perception / membrane depolarization during action potential / voltage-gated sodium channel complex / cardiac muscle cell action potential involved in contraction / Interaction between L1 and Ankyrins / voltage-gated sodium channel activity / sodium ion transport / behavioral response to pain / Phase 0 - rapid depolarisation / detection of temperature stimulus involved in sensory perception of pain ...detection of mechanical stimulus involved in sensory perception / membrane depolarization during action potential / voltage-gated sodium channel complex / cardiac muscle cell action potential involved in contraction / Interaction between L1 and Ankyrins / voltage-gated sodium channel activity / sodium ion transport / behavioral response to pain / Phase 0 - rapid depolarisation / detection of temperature stimulus involved in sensory perception of pain / calcium channel regulator activity / sodium channel regulator activity / sodium ion transmembrane transport / sensory perception of pain / post-embryonic development / response to toxic substance / Sensory perception of sweet, bitter, and umami (glutamate) taste / circadian rhythm / toxin activity / inflammatory response / axon / lipid binding / extracellular region / identical protein binding / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Arcobacter butzleri (strain RM4018) (bacteria) / Arcobacter butzleri (strain RM4018) (bacteria) /  Thrixopelma pruriens (green velvet) / Thrixopelma pruriens (green velvet) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Xu H / Rohou A | |||||||||
 Citation Citation |  Journal: Cell / Year: 2019 Journal: Cell / Year: 2019Title: Structural Basis of Nav1.7 Inhibition by a Gating-Modifier Spider Toxin. Authors: Hui Xu / Tianbo Li / Alexis Rohou / Christopher P Arthur / Foteini Tzakoniati / Evera Wong / Alberto Estevez / Christine Kugel / Yvonne Franke / Jun Chen / Claudio Ciferri / David H Hackos / ...Authors: Hui Xu / Tianbo Li / Alexis Rohou / Christopher P Arthur / Foteini Tzakoniati / Evera Wong / Alberto Estevez / Christine Kugel / Yvonne Franke / Jun Chen / Claudio Ciferri / David H Hackos / Christopher M Koth / Jian Payandeh /  Abstract: Voltage-gated sodium (Nav) channels are targets of disease mutations, toxins, and therapeutic drugs. Despite recent advances, the structural basis of voltage sensing, electromechanical coupling, and ...Voltage-gated sodium (Nav) channels are targets of disease mutations, toxins, and therapeutic drugs. Despite recent advances, the structural basis of voltage sensing, electromechanical coupling, and toxin modulation remains ill-defined. Protoxin-II (ProTx2) from the Peruvian green velvet tarantula is an inhibitor cystine-knot peptide and selective antagonist of the human Nav1.7 channel. Here, we visualize ProTx2 in complex with voltage-sensor domain II (VSD2) from Nav1.7 using X-ray crystallography and cryoelectron microscopy. Membrane partitioning orients ProTx2 for unfettered access to VSD2, where ProTx2 interrogates distinct features of the Nav1.7 receptor site. ProTx2 positions two basic residues into the extracellular vestibule to antagonize S4 gating-charge movement through an electrostatic mechanism. ProTx2 has trapped activated and deactivated states of VSD2, revealing a remarkable ∼10 Å translation of the S4 helix, providing a structural framework for activation gating in voltage-gated ion channels. Finally, our results deliver key templates to design selective Nav channel antagonists. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0342.map.gz emd_0342.map.gz | 164.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0342-v30.xml emd-0342-v30.xml emd-0342.xml emd-0342.xml | 26.4 KB 26.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0342.png emd_0342.png | 106.8 KB | ||
| Filedesc metadata |  emd-0342.cif.gz emd-0342.cif.gz | 7 KB | ||
| Others |  emd_0342_additional.map.gz emd_0342_additional.map.gz emd_0342_half_map_1.map.gz emd_0342_half_map_1.map.gz emd_0342_half_map_2.map.gz emd_0342_half_map_2.map.gz | 164.8 MB 32.6 MB 32.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0342 http://ftp.pdbj.org/pub/emdb/structures/EMD-0342 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0342 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0342 | HTTPS FTP |
-Validation report
| Summary document |  emd_0342_validation.pdf.gz emd_0342_validation.pdf.gz | 638.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_0342_full_validation.pdf.gz emd_0342_full_validation.pdf.gz | 637.8 KB | Display | |
| Data in XML |  emd_0342_validation.xml.gz emd_0342_validation.xml.gz | 15.2 KB | Display | |
| Data in CIF |  emd_0342_validation.cif.gz emd_0342_validation.cif.gz | 18 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0342 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0342 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0342 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0342 | HTTPS FTP |
-Related structure data
| Related structure data |  6n4rMC  0341C  6n4iC  6n4qC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0342.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0342.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map generated with a mask including Nav (with truncated C-terminal coiled-coil), ProTx2 and Fv fragment. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
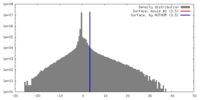
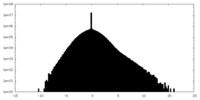
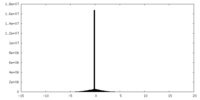
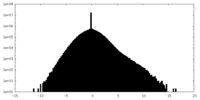
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Unsharpened map for the main map.
| File | emd_0342_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map for the main map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
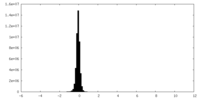
| Density Histograms |
-Half map: EM half map 2
| File | emd_0342_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
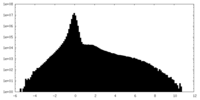
| Density Histograms |
-Half map: EM half map 1
| File | emd_0342_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
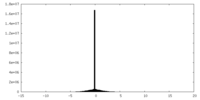
| Density Histograms |
- Sample components
Sample components
-Entire : Nav1.7 VSD2 in complex with ProTx2 and an anti-Nav Fab
| Entire | Name: Nav1.7 VSD2 in complex with ProTx2 and an anti-Nav Fab |
|---|---|
| Components |
|
-Supramolecule #1: Nav1.7 VSD2 in complex with ProTx2 and an anti-Nav Fab
| Supramolecule | Name: Nav1.7 VSD2 in complex with ProTx2 and an anti-Nav Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 245 KDa |
-Macromolecule #1: Nav1.7 VSD2-NavAb chimera
| Macromolecule | Name: Nav1.7 VSD2-NavAb chimera / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Arcobacter butzleri (strain RM4018) (bacteria) / Strain: RM4018 Arcobacter butzleri (strain RM4018) (bacteria) / Strain: RM4018 |
| Molecular weight | Theoretical: 33.453512 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MDYKDDDDKG SLVPRGSHMY LRITNIVESS FFTKFIIYLI VLNTLFMAME HHPMTEEFKN VLAIGNLVFT GIFAIEIILR IYVHRISFF KDPWSLFDSL IVTLSLVELF LADVEGLSVL RSFRLLRVFR LVTAVPQMRK IVSALISVIP GMLSVIALMT L FFYIFAIM ...String: MDYKDDDDKG SLVPRGSHMY LRITNIVESS FFTKFIIYLI VLNTLFMAME HHPMTEEFKN VLAIGNLVFT GIFAIEIILR IYVHRISFF KDPWSLFDSL IVTLSLVELF LADVEGLSVL RSFRLLRVFR LVTAVPQMRK IVSALISVIP GMLSVIALMT L FFYIFAIM ATQLFGERFP EWFGTLGESF YTLFQVMTLE SWSMGIVRPL MEVYPYAWVF FIPFIFVVTF VMINLVVAIC VD AMAILNQ KEEQHIIDEV QSHEDNINNE IIKLREEIVE LKELIKTSLK N UniProtKB: Ion transport protein, Sodium channel protein type 9 subunit alpha, Ion transport protein, Sodium channel protein type 9 subunit alpha, Ion transport protein |
-Macromolecule #2: Beta/omega-theraphotoxin-Tp2a
| Macromolecule | Name: Beta/omega-theraphotoxin-Tp2a / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thrixopelma pruriens (green velvet) Thrixopelma pruriens (green velvet) |
| Molecular weight | Theoretical: 3.839687 KDa |
| Sequence | String: YCQKWMWTCD SERKCCEGMV CRLWCKKKLW UniProtKB: Beta/omega-theraphotoxin-Tp2a |
-Macromolecule #3: Fab light chain
| Macromolecule | Name: Fab light chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.48391 KDa |
| Sequence | String: EIVLTQSPAL MAASPGEKVT ITCSVSLSIS SSNLFWYQQK SETSPKPWIY GTSKLASGVP VRFSGSGSGT SYSLTISSME AEDAATYYC QQWSSHSFTF GGGTKLEIKR ADAAPTVSIF PPSSEQLTSG GASVVCFLNN FYPKDINVKW KIDGSERQNG V LNSWTDQD ...String: EIVLTQSPAL MAASPGEKVT ITCSVSLSIS SSNLFWYQQK SETSPKPWIY GTSKLASGVP VRFSGSGSGT SYSLTISSME AEDAATYYC QQWSSHSFTF GGGTKLEIKR ADAAPTVSIF PPSSEQLTSG GASVVCFLNN FYPKDINVKW KIDGSERQNG V LNSWTDQD SKDSTYSMSS TLTLTKDEYE RHNSYTCEAT HKTSTSPIVK SFNRNEC |
-Macromolecule #4: Fab heavy chain
| Macromolecule | Name: Fab heavy chain / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.523518 KDa |
| Sequence | String: EVQLVESGGG LVKPGGSLKL SCAASGFTFS NYAMSWVRQT PEKRLEWVAT ISNGGRYTYY PDSVKGRFTI SRDNAKNSLY LQMSSLRSE DTAMYYCARH LYRYDVGGAL DYWGQGTSVT VSSAKTTAPS VYPLAPVCGD TTGSSVTLGC LVKGYFPEPV T LTWNSGSL ...String: EVQLVESGGG LVKPGGSLKL SCAASGFTFS NYAMSWVRQT PEKRLEWVAT ISNGGRYTYY PDSVKGRFTI SRDNAKNSLY LQMSSLRSE DTAMYYCARH LYRYDVGGAL DYWGQGTSVT VSSAKTTAPS VYPLAPVCGD TTGSSVTLGC LVKGYFPEPV T LTWNSGSL SSGVHTFPAV LQSDLYTLSS SVTVTSSTWP SQSITCNVAH PASSTKVDKK IEPRGPTIKP |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 10 mM Tris pH 8.0, 100 mM NaCl, 0.06% FA3, 0.1 mg/ml POPC:POPE:POPG mixed at molar ratio 3:1:1 |
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. / Pretreatment - Atmosphere: AIR / Details: Grid was coated with a thin layer of gold |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: Apply 3 uL, blot 2.5s. Ted Pella 595 filter paper.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Number grids imaged: 1 / Number real images: 25084 / Average exposure time: 10.0 sec. / Average electron dose: 41.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)