1UBD
 
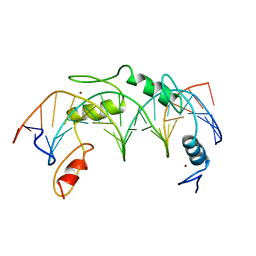 | | CO-CRYSTAL STRUCTURE OF HUMAN YY1 ZINC FINGER DOMAIN BOUND TO THE ADENO-ASSOCIATED VIRUS P5 INITIATOR ELEMENT | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*TP*CP*TP*CP*CP*AP*TP*TP*TP*TP*GP*AP*A P*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*TP*CP*AP*AP*AP*AP*TP*GP*GP*AP*GP*AP*C P*CP*CP*T)-3'), PROTEIN (YY1 ZINC FINGER DOMAIN), ... | | Authors: | Houbaviy, H.B, Usheva, A, Shenk, T, Burley, S.K. | | Deposit date: | 1996-10-04 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cocrystal structure of YY1 bound to the adeno-associated virus P5 initiator.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
2D9G
 
 | | Solution structure of the zf-RanBP domain of YY1-associated factor 2 | | Descriptor: | YY1-associated factor 2, ZINC ION | | Authors: | Zhang, H.P, Izumi, K, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-09 | | Release date: | 2006-06-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the zf-RanBP domain of YY1-associated factor 2
To be published
|
|
1ZNM
 
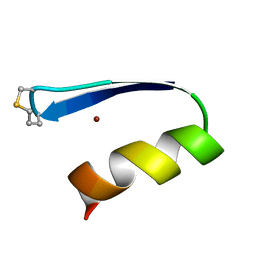 | | A zinc finger with an artificial beta-turn, original sequence taken from the third zinc finger domain of the human transcriptional repressor protein YY1 (YING and YANG 1, a delta transcription factor), nmr, 34 structures | | Descriptor: | YY1, ZINC ION | | Authors: | Viles, J.H, Patel, S.U, Mitchell, J.B.O, Moody, C.M, Justice, D.E, Uppenbrink, J, Doyle, P.M, Harris, C.J, Sadler, P.J, Thornton, J.M. | | Deposit date: | 1997-11-20 | | Release date: | 1998-04-01 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Design, synthesis and structure of a zinc finger with an artificial beta-turn.
J.Mol.Biol., 279, 1998
|
|
4C5I
 
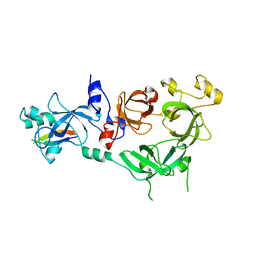 | | Crystal structure of MBTD1 YY1 complex | | Descriptor: | MBT DOMAIN-CONTAINING PROTEIN 1, TRANSCRIPTIONAL REPRESSOR PROTEIN YY1 | | Authors: | Alfieri, C, Glatt, S, Mueller, C.W. | | Deposit date: | 2013-09-12 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Structural Basis for Targeting the Chromatin Repressor Sfmbt to Polycomb Response Elements
Genes Dev., 27, 2013
|
|
2YY1
 
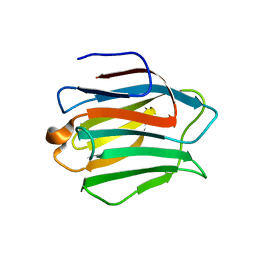 | | Crystal structure of N-terminal domain of human galectin-9 containing L-acetyllactosamine | | Descriptor: | Galectin-9, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Kishishita, S, Nishino, A, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-27 | | Release date: | 2008-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of N-terminal domain of human galectin-9 containing L-acetyllactosamine
To be Published
|
|
8YY1
 
 | | Vo domain of V/A-ATPase from Thermus thermophilus state3 | | Descriptor: | V-type ATP synthase subunit C, V-type ATP synthase subunit I, V-type ATP synthase, ... | | Authors: | Nishida, Y, Kishikawa, J, Nakano, A, Yokoyama, K. | | Deposit date: | 2024-04-03 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Rotary mechanism of the prokaryotic V o motor driven by proton motive force.
Nat Commun, 15, 2024
|
|
7YY1
 
 | |
4YY1
 
 | | The structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013) in complex with human receptor analog 6'SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, ... | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013)
To Be Published
|
|
6YY1
 
 | | Arabidopsis aspartate transcarbamoylase in apo state | | Descriptor: | GLYCEROL, PYRB | | Authors: | Ramon Maiques, S, Del Cano Ochoa, F, Bellin, L, Mohlmann, T. | | Deposit date: | 2020-05-04 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Mechanisms of feedback inhibition and sequential firing of active sites in plant aspartate transcarbamoylase.
Nat Commun, 12, 2021
|
|
5YY1
 
 | | Co-crystal Structure of KRAS (G12C) covalently bound with Quinazoline based inhibitor JBI739 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[6-chloranyl-8-fluoranyl-7-[2-(trifluoromethyl)phenyl]quinazolin-4-yl]piperazin-1-yl]propan-1-one, GTPase KRas, ... | | Authors: | Swaminathan, S, Thakur, M.K, Kandan, S, Gautam, A, Kanavalli, M, Simhadri, P, Gosu, R. | | Deposit date: | 2017-12-07 | | Release date: | 2018-04-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Co-crystal Structure of KRAS (G12C) covalently bound with Quinazoline based inhibitor JBI739
To Be Published
|
|
1YY1
 
 | |
1YY2
 
 | |
1L9G
 
 | | CRYSTAL STRUCTURE OF URACIL-DNA GLYCOSYLASE FROM T. MARITIMA | | Descriptor: | Conserved hypothetical protein, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Rajashankar, K.R, Dodatko, T, Thirumuruhan, R.A, Sandigursky, M, Bresnik, A, Chance, M.R, Franklin, W.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-03-22 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of uracil-DNA glycosylase from T. Maritima
To be Published
|
|
1VK2
 
 | |
1TTU
 
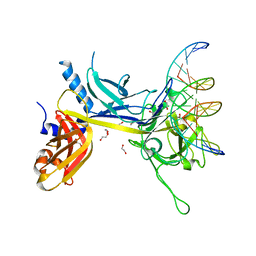 | | Crystal Structure of CSL bound to DNA | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*T)-3', 5'-D(*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3', ... | | Authors: | Kovall, R.A, Hendrickson, W.A. | | Deposit date: | 2004-06-23 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA
Embo J., 23, 2004
|
|
4YY0
 
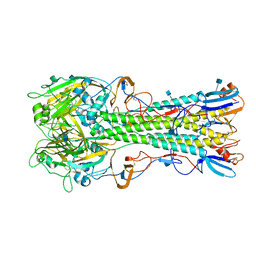 | | The structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, HA2 | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013)
To Be Published
|
|
3IXS
 
 | | Ring1B C-terminal domain/RYBP C-terminal domain Complex | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, E3 ubiquitin-protein ligase RING2, ... | | Authors: | Wang, R, Taylor, A.B, Kim, C.A. | | Deposit date: | 2009-09-04 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Polycomb Group Targeting through Different Binding Partners of RING1B C-Terminal Domain.
Structure, 18, 2010
|
|
4C5G
 
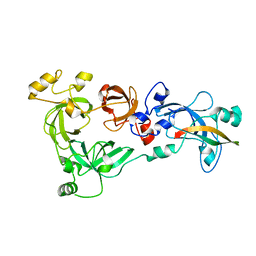 | |
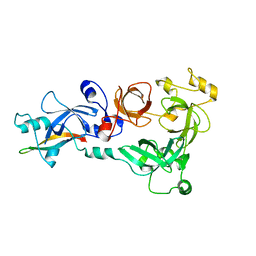
4C5H
 
 | |
4C5E
 
 | |
8PP6
 
 | | human RYBP-PRC1 bound to H2AK118ub1 nucleosome | | Descriptor: | DNA (215-MER), Histone H2A, Histone H2B, ... | | Authors: | Ciapponi, M, Benda, C, Mueller, J. | | Deposit date: | 2023-07-06 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis of the histone ubiquitination read-write mechanism of RYBP-PRC1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
9DBY
 
 | | ncPRC1RYBP bound to singly modified H2AK119Ub nucleosome | | Descriptor: | DNA (187-MER), E3 ubiquitin-protein ligase RING2, Histone H2A type 1, ... | | Authors: | Godinez-Lopez, V, Valencia-Sanchez, M.I, Armache, J.P, Armache, K.-J. | | Deposit date: | 2024-08-24 | | Release date: | 2024-11-20 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Read-write mechanisms of H2A ubiquitination by Polycomb repressive complex 1.
Nature, 636, 2024
|
|
9DDE
 
 | | ncPRC1RYBP bound to H2AK119Ub/H1.4 chromatosome | | Descriptor: | DNA (187-MER), E3 ubiquitin-protein ligase RING2, Histone H1.4, ... | | Authors: | Godinez-Lopez, V, Valencia-Sanchez, M.I, Armache, J.P, Armache, K.-J. | | Deposit date: | 2024-08-28 | | Release date: | 2024-11-20 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Read-write mechanisms of H2A ubiquitination by Polycomb repressive complex 1.
Nature, 636, 2024
|
|
9DG3
 
 | | ncPRC1RYBP Delta-linker mutant bound to singly modified H2AK119Ub nucleosome | | Descriptor: | DNA (187-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Godinez-Lopez, V, Valencia-Sanchez, M.I, Armache, J.P, Armache, K.-J. | | Deposit date: | 2024-09-01 | | Release date: | 2024-11-20 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Read-write mechanisms of H2A ubiquitination by Polycomb repressive complex 1.
Nature, 636, 2024
|
|
4WRH
 
 | | AKR1C3 complexed with breakdown product of N-(tert-butyl)-2-(2-chloro-4-(((3-mercapto-5-methyl-4H-1,2,4-triazol-4-yl)amino)methyl)-6-methoxyphenoxy)acetamide | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(aminomethyl)-2-chloro-6-methoxyphenoxy]-N-tert-butylacetamide, 5-methyl-4H-1,2,4-triazole-3-thiol, ... | | Authors: | Squire, C.J, Flanagan, J.U, Yosaatmadja, Y. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Breakdown product of N-(tert-butyl)-2-(2-chloro-4-(((3-mercapto-5-methyl-4H-1,2,4-triazol-4-yl)amino)methyl)-6-methoxyphenoxy)acetamide trapped in active site of AKR1C3
To Be Published
|
|
