8S7L
 
 | | Crystal structure of a double mutant of VirB8-like OrfG central and C-terminal domains of Streptococcus thermophilus ICESt3 (Gram positive conjugative type IV secretion system). | | Descriptor: | Putative transfer protein | | Authors: | Favier, F, Didierjean, C, Maffo-Woulefack, R, Douzi, B, Leblond-Bourget, N. | | Deposit date: | 2024-03-03 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Elucidating assembly and function of VirB8 cell wall subunits refines the DNA translocation model in Gram-positive T4SSs.
Sci Adv, 11, 2025
|
|
3GMC
 
 | | Crystal Structure of 2-Methyl-3-hydroxypyridine-5-carboxylic acid Oxygenase with substrate bound | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxy-6-methylpyridine-3-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | McCulloch, K.M, Mukherjee, T, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-03-13 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the PLP degradative enzyme 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase from Mesorhizobium loti MAFF303099 and its mechanistic implications.
Biochemistry, 48, 2009
|
|
3GMB
 
 | | Crystal Structure of 2-Methyl-3-hydroxypyridine-5-carboxylic acid Oxygenase | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | McCulloch, K.M, Mukherjee, T, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-03-13 | | Release date: | 2009-04-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the PLP degradative enzyme 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase from Mesorhizobium loti MAFF303099 and its mechanistic implications.
Biochemistry, 48, 2009
|
|
7PKW
 
 | | Crystal structure of VIRB8-like OrfG central and C-terminal domains of Streptococcus thermophilus ICESt3 (Gram positive conjugative type IV secretion system). | | Descriptor: | GLYCEROL, Putative transfer protein, SULFATE ION | | Authors: | Favier, F, Didierjean, C, Cappele, J, Douzi, B, Leblond-Bourget, N. | | Deposit date: | 2021-08-27 | | Release date: | 2022-09-07 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.835 Å) | | Cite: | Elucidating assembly and function of VirB8 cell wall subunits refines the DNA translocation model in Gram-positive T4SSs.
Sci Adv, 11, 2025
|
|
4BAY
 
 | | Phosphomimetic mutant of LSD1-8a splicing variant in complex with CoREST | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1 | | Authors: | Toffolo, E, Paganini, L, Rusconi, F, Tortorici, M, Pilotto, S, Verpelli, C, Tedeschi, G, Maffioli, E, Sala, C, Mattevi, A, Battaglioli, E. | | Deposit date: | 2012-09-17 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Phosphorylation of Neuronal Lysine-Specific Demethylase 1Lsd1/Kdm1A Impairs Transcriptional Repression by Regulating Interaction with Corest and Histone Deacetylases Hdac1/2.
J.Neurochem., 128, 2014
|
|
7QPG
 
 | | Human RZZ kinetochore corona complex. | | Descriptor: | Centromere/kinetochore protein zw10 homolog, Kinetochore-associated protein 1, Protein zwilch homolog | | Authors: | Raisch, T, Ciossani, G, d'Amico, E, Cmetowski, V, Carmignani, S, Maffini, S, Merino, F, Wohlgemuth, S, Vetter, I.R, Raunser, S, Musacchio, A. | | Deposit date: | 2022-01-04 | | Release date: | 2022-03-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the RZZ complex and molecular basis of Spindly-driven corona assembly at human kinetochores.
Embo J., 41, 2022
|
|
2Z7B
 
 | | Crystal Structure of Mesorhizobium loti 3-hydroxy-2-methylpyridine-4,5-dicarboxylate decarboxylase | | Descriptor: | MANGANESE (II) ION, Mlr6791 protein | | Authors: | McCulloch, K.M, Mukherjee, T, Ealick, S.E, Begley, T.P. | | Deposit date: | 2007-08-17 | | Release date: | 2007-11-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Gene Identification and Structural Characterization of the Pyridoxal 5'-Phosphate Degradative Protein 3-Hydroxy-2-methylpyridine-4,5-dicarboxylate Decarboxylase from Mesorhizobium loti MAFF303099
Biochemistry, 46, 2007
|
|
2MH5
 
 | | Structure and NMR assignments of lantibiotic NAI-107 in DPC micelles | | Descriptor: | Lantibiotic 107891, dodecyl 2-(trimethylammonio)ethyl phosphate | | Authors: | Munch, D, Muller, A, Schneider, T, Kohl, B, Wenzel, M, Bandow, J, Maffioli, S, Sosio, M, Donadio, S, Wimmer, R, Sahl, H. | | Deposit date: | 2013-11-18 | | Release date: | 2014-03-05 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | The Lantibiotic NAI-107 Binds to Bactoprenol-bound Cell Wall Precursors and Impairs Membrane Functions.
J.Biol.Chem., 289, 2014
|
|
1T1V
 
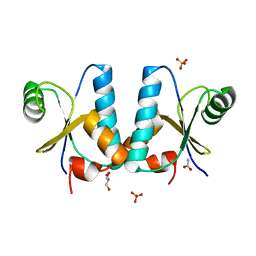 | | Crystal Structure of the Glutaredoxin-like Protein SH3BGRL3 at 1.6 A resolution | | Descriptor: | ACETIC ACID, GLYCEROL, SH3 domain-binding glutamic acid-rich protein-like 3, ... | | Authors: | Nardini, M, Mazzocco, M, Massaro, M, Maffei, M, Vergano, A, Donadini, A, Scartezzini, M, Bolognesi, M. | | Deposit date: | 2004-04-19 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the glutaredoxin-like protein SH3BGRL3 at 1.6 A resolution
Biochem.Biophys.Res.Commun., 318, 2004
|
|
5X21
 
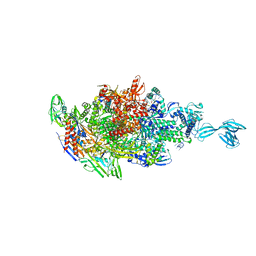 | | Crystal structure of Thermus thermophilus transcription initiation complex with GpA and pseudouridimycin (PUM) | | Descriptor: | (1S)-1,4-anhydro-5-[(N-carbamimidoylglycyl-N~2~-hydroxy-L-glutaminyl)amino]-5-deoxy-1-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)-D-ribitol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Zhang, Y, Ebright, R. | | Deposit date: | 2017-01-29 | | Release date: | 2017-07-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.323 Å) | | Cite: | Antibacterial Nucleoside-Analog Inhibitor of Bacterial RNA Polymerase.
Cell, 169, 2017
|
|
5X22
 
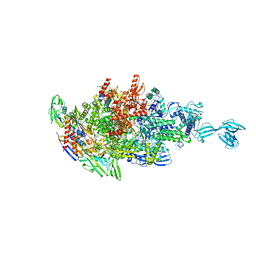 | |
8EVM
 
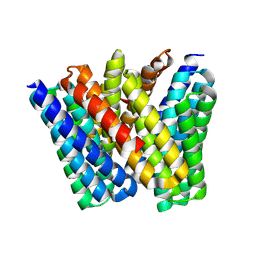 | |
7UNH
 
 | |
7UNI
 
 | | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester, SP2-ZnPPaM | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, SP2-ZnPPaM designed chlorophyll dimer protein, ... | | Authors: | Kennedy, M.A, Stoddard, B.L, Ennist, N.M. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 2024
|
|
7UNJ
 
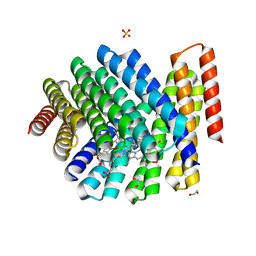 | | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester matching geometry of purple bacterial special pair, SP1-ZnPPaM | | Descriptor: | 1,2-ETHANEDIOL, SP1-ZnPPaM designed chlorophyll dimer protein, SULFATE ION, ... | | Authors: | Kennedy, M.A, Stoddard, B.L, Ennist, N.M. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 2024
|
|
8ARF
 
 | |
6EQT
 
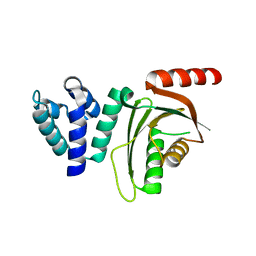 | |
6C0W
 
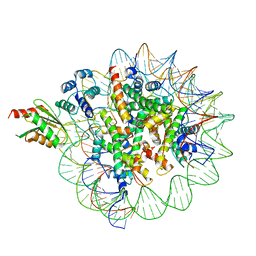 | | Cryo-EM structure of human kinetochore protein CENP-N with the centromeric nucleosome containing CENP-A | | Descriptor: | 147 mer DNA, Centromere protein N, Histone H2A, ... | | Authors: | Zhou, K, Pentakota, S, Vetter, I.R, Morgan, G.P, Petrovic, A, Musacchio, A, Luger, K. | | Deposit date: | 2018-01-02 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Decoding the centromeric nucleosome through CENP-N.
Elife, 6, 2017
|
|
7OK3
 
 | |
7OK4
 
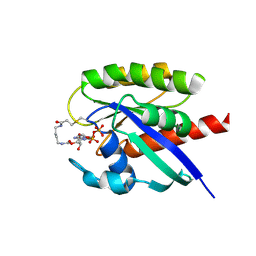 | |
2Z9W
 
 | | Crystal structure of pyridoxamine-pyruvate aminotransferase complexed with pyridoxal | | Descriptor: | 3-HYDROXY-5-(HYDROXYMETHYL)-2-METHYLISONICOTINALDEHYDE, Aspartate aminotransferase, GLYCEROL, ... | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
2Z9U
 
 | | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti at 2.0 A resolution | | Descriptor: | Aspartate aminotransferase, GLYCEROL, SULFATE ION | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
2Z9V
 
 | | Crystal structure of pyridoxamine-pyruvate aminotransferase complexed with pyridoxamine | | Descriptor: | 4-(AMINOMETHYL)-5-(HYDROXYMETHYL)-2-METHYLPYRIDIN-3-OL, Aspartate aminotransferase, GLYCEROL, ... | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
2Z9X
 
 | | Crystal structure of pyridoxamine-pyruvate aminotransferase complexed with pyridoxyl-L-alanine | | Descriptor: | 3-HYDROXY-5-(HYDROXYMETHYL)-2-METHYLISONICOTINALDEHYDE, ALANINE, Aspartate aminotransferase, ... | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
5UAO
 
 | | Crystal structure of MibH, a lathipeptide tryptophan 5-halogenase | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Tryptophane-5-halogenase | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Two Flavoenzymes Catalyze the Post-Translational Generation of 5-Chlorotryptophan and 2-Aminovinyl-Cysteine during NAI-107 Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
