6WKR
 
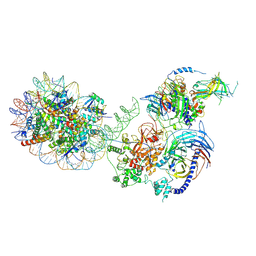 | | PRC2-AEBP2-JARID2 bound to H2AK119ub1 nucleosome | | Descriptor: | DNA (314-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Kasinath, V, Nogales, E, Beck, C, Sauer, P, Poepsel, S, Kosmatka, J, Faini, M, Toso, D, Aebersold, R. | | Deposit date: | 2020-04-16 | | Release date: | 2021-02-03 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | JARID2 and AEBP2 regulate PRC2 in the presence of H2AK119ub1 and other histone modifications.
Science, 371, 2021
|
|
3GGZ
 
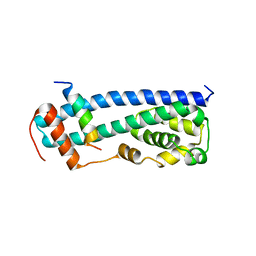 | |
5H7P
 
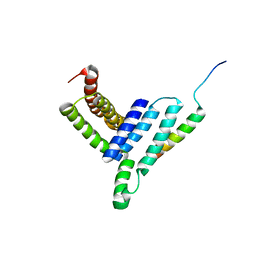 | | NMR structure of the Vta1NTD-Did2(176-204) complex | | Descriptor: | Vacuolar protein sorting-associated protein VTA1, Vacuolar protein-sorting-associated protein 46 | | Authors: | Shen, J, Yang, Z, Wild, C.J. | | Deposit date: | 2016-11-20 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR studies on the interactions between yeast Vta1 and Did2 during the multivesicular bodies sorting pathway
Sci Rep, 6, 2016
|
|
6C23
 
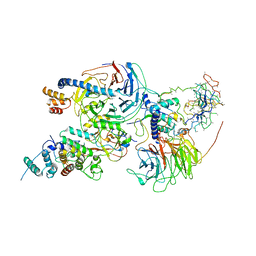 | | Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Compact Active State | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, JARID2-substrate, ... | | Authors: | Kasinath, V, Faini, M, Poepsel, S, Reif, D, Feng, A, Stjepanovic, G, Aebersold, R, Nogales, E. | | Deposit date: | 2018-01-05 | | Release date: | 2018-01-24 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of human PRC2 with its cofactors AEBP2 and JARID2.
Science, 359, 2018
|
|
6C24
 
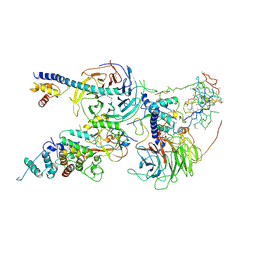 | | Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Extended Active State | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, JARID2-substrate, ... | | Authors: | Kasinath, V, Faini, M, Poepsel, S, Reif, D, Feng, A, Stjepanovic, G, Aebersold, R, Nogales, E. | | Deposit date: | 2018-01-06 | | Release date: | 2018-01-24 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of human PRC2 with its cofactors AEBP2 and JARID2.
Science, 359, 2018
|
|
4X3E
 
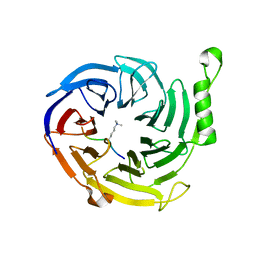 | |
7QGF
 
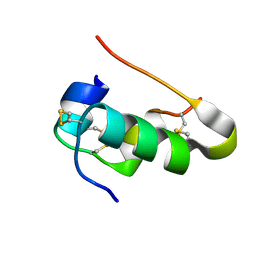 | | Cubic Insulin SAD phasing at 14.2 keV | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Nanao, M.H, Basu, S. | | Deposit date: | 2021-12-08 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.203 Å) | | Cite: | ID23-2: an automated and high-performance microfocus beamline for macromolecular crystallography at the ESRF.
J.Synchrotron Radiat., 29, 2022
|
|
7QRZ
 
 | |
4AYA
 
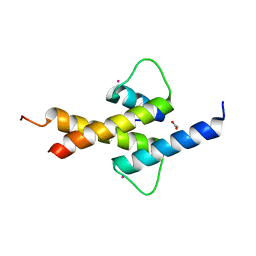 | | Crystal structure of ID2 HLH homodimer at 2.1A resolution | | Descriptor: | ACETATE ION, DNA-BINDING PROTEIN INHIBITOR ID-2, POTASSIUM ION | | Authors: | Wong, M.V, Jiang, S, Palasingam, P, Kolatkar, P.R. | | Deposit date: | 2012-06-19 | | Release date: | 2012-11-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | A Divalent Ion is Crucial in the Structure and Dominant-Negative Function of Id Proteins, a Class of Helix-Loop-Helix Transcription Regulators.
Plos One, 7, 2012
|
|
3GGY
 
 | | Crystal Structure of S.cerevisiae Ist1 N-terminal domain | | Descriptor: | Increased sodium tolerance protein 1 | | Authors: | Xiao, J, Xu, Z. | | Deposit date: | 2009-03-02 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of Ist1 function and Ist1-Did2 interaction in the multivesicular body pathway and cytokinesis.
MOLECULAR BIOLOGY OF THE CELL, 20, 2009
|
|
2RQ5
 
 | |
3T5X
 
 | | PCID2:DSS1 Structure | | Descriptor: | 26S proteasome complex subunit DSS1, PCI domain-containing protein 2 | | Authors: | Stewart, M, Ellisdon, A.M. | | Deposit date: | 2011-07-28 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.123 Å) | | Cite: | Structural basis for the assembly and nucleic acid binding of the TREX-2 transcription-export complex.
Nat.Struct.Mol.Biol., 19, 2012
|
|
7AUX
 
 | | Crystal structure of OXA-48 beta-lactamase in the complex with the inhbitor ID2 | | Descriptor: | 6-(4-carboxyphenyl)-3-(4-ethylphenyl)-2~{H}-pyrazolo[3,4-b]pyridine-4-carboxylic acid, Beta-lactamase, CHLORIDE ION | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Garofalo, B, Ombrato, R. | | Deposit date: | 2020-11-03 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Novel Chemical Series of OXA-48 beta-Lactamase Inhibitors by High-Throughput Screening.
Pharmaceuticals, 14, 2021
|
|
8DOV
 
 | | Crystal structure of the Shr Hemoglobin Interacting Domain 2 (HID2) in complex with Hemoglobin | | Descriptor: | GLYCEROL, Heme-binding protein Shr, Hemoglobin subunit alpha, ... | | Authors: | Macdonald, R, Mahoney, B.J, Cascio, D, Clubb, R.T. | | Deposit date: | 2022-07-14 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Shr receptor from Streptococcus pyogenes uses a cap and release mechanism to acquire heme-iron from human hemoglobin.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
9D2H
 
 | |
6NQ3
 
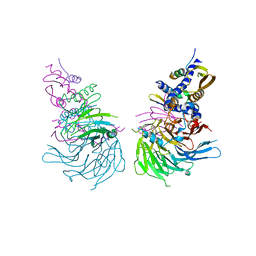 | | Crystal Structure of a SUZ12-RBBP4-PHF19-JARID2 Heterotetrameric Complex | | Descriptor: | Histone-binding protein RBBP4, PHD finger protein 19, Polycomb protein SUZ12, ... | | Authors: | Chen, S, Jiao, L, Liu, X. | | Deposit date: | 2019-01-19 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | A Dimeric Structural Scaffold for PRC2-PCL Targeting to CpG Island Chromatin.
Mol.Cell, 77, 2020
|
|
8G01
 
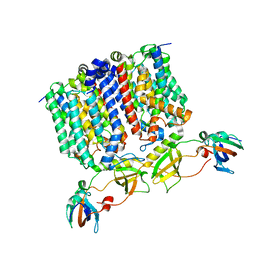 | | YES Complex - E. coli MraY, Protein E ID21, E. coli SlyD | | Descriptor: | FKBP-type peptidyl-prolyl cis-trans isomerase SlyD, GPE, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Orta, A.K, Clemons, W.M, Riera, N. | | Deposit date: | 2023-01-31 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The mechanism of the phage-encoded protein antibiotic from Phi X174.
Science, 381, 2023
|
|
5WAI
 
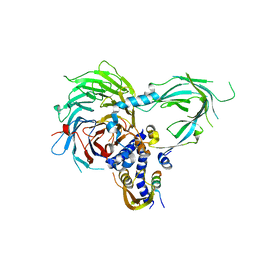 | | Crystal Structure of a Suz12-Rbbp4-Jarid2-Aebp2 Heterotetrameric Complex | | Descriptor: | Histone-binding protein RBBP4, Jumonji, AT-rich interactive domain 2, ... | | Authors: | Chen, S, Jiao, L, Liu, X. | | Deposit date: | 2017-06-26 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Unique Structural Platforms of Suz12 Dictate Distinct Classes of PRC2 for Chromatin Binding.
Mol. Cell, 69, 2018
|
|
7VDV
 
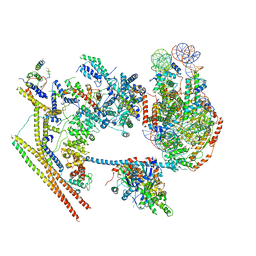 | | The overall structure of human chromatin remodeling PBAF-nucleosome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AT-rich interactive domain-containing protein 2,AT-rich interactive domain-containing protein 2, Actin, ... | | Authors: | Chen, Z.C, Chen, K.J, Yuan, J.J. | | Deposit date: | 2021-09-07 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of human chromatin-remodelling PBAF complex bound to a nucleosome.
Nature, 605, 2022
|
|
8VML
 
 | | PRC2_AJ1-450 bound to H3K4me3 | | Descriptor: | AEPB2, EED, EZH2, ... | | Authors: | Cookis, T, Nogales, E. | | Deposit date: | 2024-01-13 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the inhibition of PRC2 by active transcription histone posttranslational modifications.
Nat.Struct.Mol.Biol., 32, 2025
|
|
9FUP
 
 | | Serial microseconds crystallography at ID29 using fixed-target (small foils): A2a adenosine receptor co-crystallised with Istradefylline | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 8-[(~{E})-2-(3,4-dimethoxyphenyl)ethenyl]-1,3-diethyl-7-methyl-purine-2,6-dione, ... | | Authors: | Orlans, J, Rose, S.L, Ferguson, G, Lebon, G, Basu, S, de Sanctis, D. | | Deposit date: | 2024-06-26 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Advancing macromolecular structure determination with microsecond X-ray pulses at a 4th generation synchrotron.
Commun Chem, 8, 2025
|
|
9FU1
 
 | | Serial microseconds crystallography at ID29 using fixed-target (large foils): Proteinase K with 50 um spacing between X-ray pulses | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Orlans, J, Rose, S.L, Basu, S, de Sanctis, D. | | Deposit date: | 2024-06-25 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Advancing macromolecular structure determination with microsecond X-ray pulses at a 4th generation synchrotron.
Commun Chem, 8, 2025
|
|
9FTY
 
 | | Serial microseconds crystallography at ID29 using fixed-target (small foils): Proteinase K with 20 um spacing between X-ray pulses | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Orlans, J, Rose, S.L, Basu, S, de Sanctis, D. | | Deposit date: | 2024-06-25 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Advancing macromolecular structure determination with microsecond X-ray pulses at a 4th generation synchrotron.
Commun Chem, 8, 2025
|
|
9FTS
 
 | | Serial microseconds crystallography at ID29 using SACLA extruder: Thaumatin | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Orlans, J, Rose, S.L, Basu, S, de Sanctis, D. | | Deposit date: | 2024-06-25 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Advancing macromolecular structure determination with microsecond X-ray pulses at a 4th generation synchrotron.
Commun Chem, 8, 2025
|
|
9FTU
 
 | | Serial microseconds crystallography at ID29 using ASU extruder: Thaumatin | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Orlans, J, Rose, S.L, Basu, S, de Sanctis, D. | | Deposit date: | 2024-06-25 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Advancing macromolecular structure determination with microsecond X-ray pulses at a 4th generation synchrotron.
Commun Chem, 8, 2025
|
|
