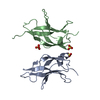
Entry Database : PDB / ID : 3jv4Title Crystal structure of the dimerization domains p50 and RelB Nuclear factor NF-kappa-B p105 subunit Transcription factor RelB Keywords / / / / / / / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Mus musculus (house mouse)Method / / Resolution : 3.15 Å Authors Vu, D. / Huang, D.B. / Ghosh, G. Journal : J.Mol.Biol. / Year : 2013Title : A structural basis for selective dimerization by NF-kappa B RelB.Authors : Vu, D. / Huang, D.B. / Vemu, A. / Ghosh, G. History Deposition Sep 15, 2009 Deposition site / Processing site Revision 1.0 Nov 24, 2010 Provider / Type Revision 1.1 Jul 13, 2011 Group Revision 1.2 Mar 27, 2013 Group Revision 1.3 Sep 4, 2013 Group Revision 1.4 Feb 21, 2024 Group / Database references / Refinement descriptionCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / struct_ncs_dom_lim Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_ncs_dom_lim.beg_auth_comp_id / _struct_ncs_dom_lim.beg_label_comp_id / _struct_ncs_dom_lim.beg_label_seq_id / _struct_ncs_dom_lim.end_auth_comp_id / _struct_ncs_dom_lim.end_label_comp_id / _struct_ncs_dom_lim.end_label_seq_id
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords TRANSCRIPTION /
TRANSCRIPTION /  NF-kB protein /
NF-kB protein /  Heterodimer / RelB and p50 / Activator /
Heterodimer / RelB and p50 / Activator /  Nucleus /
Nucleus /  Phosphoprotein /
Phosphoprotein /  Transcription regulation / ANK repeat /
Transcription regulation / ANK repeat /  Apoptosis / DNA-binding /
Apoptosis / DNA-binding /  S-nitrosylation
S-nitrosylation Function and homology information
Function and homology information actinin binding / cellular response to angiotensin / negative regulation of cytokine production / positive regulation of miRNA metabolic process / T-helper 1 type immune response / non-canonical NF-kappaB signal transduction / cellular response to cytokine stimulus / cellular response to organic cyclic compound / antigen processing and presentation / positive regulation of transcription initiation by RNA polymerase II / canonical NF-kappaB signal transduction / lymph node development / cellular response to interleukin-1 / JNK cascade / cellular response to brain-derived neurotrophic factor stimulus /
actinin binding / cellular response to angiotensin / negative regulation of cytokine production / positive regulation of miRNA metabolic process / T-helper 1 type immune response / non-canonical NF-kappaB signal transduction / cellular response to cytokine stimulus / cellular response to organic cyclic compound / antigen processing and presentation / positive regulation of transcription initiation by RNA polymerase II / canonical NF-kappaB signal transduction / lymph node development / cellular response to interleukin-1 / JNK cascade / cellular response to brain-derived neurotrophic factor stimulus /  heat shock protein binding / response to muscle stretch / transcription repressor complex / Neutrophil degranulation / protein sequestering activity / response to cytokine /
heat shock protein binding / response to muscle stretch / transcription repressor complex / Neutrophil degranulation / protein sequestering activity / response to cytokine /  transcription coregulator activity / RNA polymerase II transcription regulatory region sequence-specific DNA binding / B cell receptor signaling pathway / circadian regulation of gene expression / response to organic cyclic compound / cellular response to virus / DNA-binding transcription repressor activity, RNA polymerase II-specific / negative regulation of inflammatory response / cellular response to mechanical stimulus / cellular response to nicotine / positive regulation of canonical Wnt signaling pathway /
transcription coregulator activity / RNA polymerase II transcription regulatory region sequence-specific DNA binding / B cell receptor signaling pathway / circadian regulation of gene expression / response to organic cyclic compound / cellular response to virus / DNA-binding transcription repressor activity, RNA polymerase II-specific / negative regulation of inflammatory response / cellular response to mechanical stimulus / cellular response to nicotine / positive regulation of canonical Wnt signaling pathway /  MAPK cascade / sequence-specific double-stranded DNA binding /
MAPK cascade / sequence-specific double-stranded DNA binding /  gene expression / cellular response to tumor necrosis factor /
gene expression / cellular response to tumor necrosis factor /  double-stranded DNA binding / DNA-binding transcription activator activity, RNA polymerase II-specific / response to oxidative stress / cellular response to lipopolysaccharide /
double-stranded DNA binding / DNA-binding transcription activator activity, RNA polymerase II-specific / response to oxidative stress / cellular response to lipopolysaccharide /  transcription regulator complex / sequence-specific DNA binding / transcription cis-regulatory region binding / DNA-binding transcription factor activity, RNA polymerase II-specific /
transcription regulator complex / sequence-specific DNA binding / transcription cis-regulatory region binding / DNA-binding transcription factor activity, RNA polymerase II-specific /  inflammatory response / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity / negative regulation of gene expression /
inflammatory response / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity / negative regulation of gene expression /  innate immune response / negative regulation of DNA-templated transcription /
innate immune response / negative regulation of DNA-templated transcription /  centrosome /
centrosome /  synapse / apoptotic process /
synapse / apoptotic process /  chromatin binding /
chromatin binding /  chromatin / protein-containing complex binding / positive regulation of gene expression / regulation of transcription by RNA polymerase II / negative regulation of apoptotic process /
chromatin / protein-containing complex binding / positive regulation of gene expression / regulation of transcription by RNA polymerase II / negative regulation of apoptotic process /  protein kinase binding / positive regulation of DNA-templated transcription / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / protein-containing complex /
protein kinase binding / positive regulation of DNA-templated transcription / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / protein-containing complex /  mitochondrion /
mitochondrion /  DNA binding
DNA binding
 Mus musculus (house mouse)
Mus musculus (house mouse) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 3.15 Å
MOLECULAR REPLACEMENT / Resolution: 3.15 Å  Authors
Authors Citation
Citation Journal: J.Mol.Biol. / Year: 2013
Journal: J.Mol.Biol. / Year: 2013 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 3jv4.cif.gz
3jv4.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb3jv4.ent.gz
pdb3jv4.ent.gz PDB format
PDB format 3jv4.json.gz
3jv4.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/jv/3jv4
https://data.pdbj.org/pub/pdb/validation_reports/jv/3jv4 ftp://data.pdbj.org/pub/pdb/validation_reports/jv/3jv4
ftp://data.pdbj.org/pub/pdb/validation_reports/jv/3jv4 Links
Links Assembly
Assembly



 Components
Components
 Mus musculus (house mouse) / Gene: RelB / Plasmid: PET15b / Production host:
Mus musculus (house mouse) / Gene: RelB / Plasmid: PET15b / Production host: 
 Escherichia coli (E. coli) / Strain (production host): BL21 / References: UniProt: Q04863
Escherichia coli (E. coli) / Strain (production host): BL21 / References: UniProt: Q04863
 Mus musculus (house mouse) / Gene: p50 / Plasmid: PET29b / Production host:
Mus musculus (house mouse) / Gene: p50 / Plasmid: PET29b / Production host: 
 Escherichia coli (E. coli) / Strain (production host): BL21 / References: UniProt: P25799
Escherichia coli (E. coli) / Strain (production host): BL21 / References: UniProt: P25799 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation
 ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 Å
ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 Å : 1.5418 Å / Relative weight: 1
: 1.5418 Å / Relative weight: 1  Processing
Processing :
:  MOLECULAR REPLACEMENT / Resolution: 3.15→29.76 Å / Rfactor Rfree error: 0.011 / Occupancy max: 1 / Occupancy min: 1 / FOM work R set: 0.836 / Data cutoff high absF: 32597 / Data cutoff low absF: 0 / Isotropic thermal model: GROUP / Cross valid method: THROUGHOUT / σ(F): 2 / Stereochemistry target values: Engh & Huber
MOLECULAR REPLACEMENT / Resolution: 3.15→29.76 Å / Rfactor Rfree error: 0.011 / Occupancy max: 1 / Occupancy min: 1 / FOM work R set: 0.836 / Data cutoff high absF: 32597 / Data cutoff low absF: 0 / Isotropic thermal model: GROUP / Cross valid method: THROUGHOUT / σ(F): 2 / Stereochemistry target values: Engh & Huber Movie
Movie Controller
Controller













 PDBj
PDBj





















