[English] 日本語
 Yorodumi
Yorodumi- PDB-2ktb: Solution Structure of the Second Bromodomain of Human Polybromo i... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2ktb | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Solution Structure of the Second Bromodomain of Human Polybromo in complex with an acetylated peptide from Histone 3 | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords |  PROTEIN BINDING / PROTEIN BINDING /  bromodomain / bromodomain /  Alternative splicing / Chromatin regulator / DNA-binding / Alternative splicing / Chromatin regulator / DNA-binding /  Nucleus / Nucleus /  Phosphoprotein / Phosphoprotein /  Transcription / Transcription /  Transcription regulation Transcription regulation | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of G0 to G1 transition / regulation of nucleotide-excision repair / RSC-type complex /  SWI/SNF complex / regulation of mitotic metaphase/anaphase transition / positive regulation of double-strand break repair / positive regulation of T cell differentiation / nuclear chromosome / RUNX1 interacts with co-factors whose precise effect on RUNX1 targets is not known / regulation of G1/S transition of mitotic cell cycle ...regulation of G0 to G1 transition / regulation of nucleotide-excision repair / RSC-type complex / SWI/SNF complex / regulation of mitotic metaphase/anaphase transition / positive regulation of double-strand break repair / positive regulation of T cell differentiation / nuclear chromosome / RUNX1 interacts with co-factors whose precise effect on RUNX1 targets is not known / regulation of G1/S transition of mitotic cell cycle ...regulation of G0 to G1 transition / regulation of nucleotide-excision repair / RSC-type complex /  SWI/SNF complex / regulation of mitotic metaphase/anaphase transition / positive regulation of double-strand break repair / positive regulation of T cell differentiation / nuclear chromosome / RUNX1 interacts with co-factors whose precise effect on RUNX1 targets is not known / regulation of G1/S transition of mitotic cell cycle / positive regulation of myoblast differentiation / transcription elongation by RNA polymerase II / positive regulation of cell differentiation / SWI/SNF complex / regulation of mitotic metaphase/anaphase transition / positive regulation of double-strand break repair / positive regulation of T cell differentiation / nuclear chromosome / RUNX1 interacts with co-factors whose precise effect on RUNX1 targets is not known / regulation of G1/S transition of mitotic cell cycle / positive regulation of myoblast differentiation / transcription elongation by RNA polymerase II / positive regulation of cell differentiation /  kinetochore / RMTs methylate histone arginines / kinetochore / RMTs methylate histone arginines /  nuclear matrix / mitotic cell cycle / nuclear matrix / mitotic cell cycle /  chromatin remodeling / negative regulation of cell population proliferation / chromatin remodeling / negative regulation of cell population proliferation /  chromatin binding / chromatin binding /  chromatin / regulation of transcription by RNA polymerase II / chromatin / regulation of transcription by RNA polymerase II /  DNA binding / DNA binding /  nucleoplasm / nucleoplasm /  nucleus nucleusSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  SOLUTION NMR / SOLUTION NMR /  simulated annealing, torsion angle dynamics simulated annealing, torsion angle dynamics | |||||||||
 Authors Authors | Charlop-Powers, Z. / Zhang, Q. / Zeng, L. | |||||||||
 Citation Citation |  Journal: Cell Res. / Year: 2010 Journal: Cell Res. / Year: 2010Title: Structural insights into selective histone H3 recognition by the human Polybromo bromodomain 2. Authors: Charlop-Powers, Z. / Zeng, L. / Zhang, Q. / Zhou, M.M. | |||||||||
| History |
|
- Structure visualization
Structure visualization
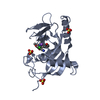
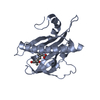
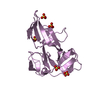
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2ktb.cif.gz 2ktb.cif.gz | 885.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2ktb.ent.gz pdb2ktb.ent.gz | 740.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2ktb.json.gz 2ktb.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/kt/2ktb https://data.pdbj.org/pub/pdb/validation_reports/kt/2ktb ftp://data.pdbj.org/pub/pdb/validation_reports/kt/2ktb ftp://data.pdbj.org/pub/pdb/validation_reports/kt/2ktb | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 2231.602 Da / Num. of mol.: 1 / Fragment: H3(1-20)K14ac / Mutation: K14(ALY) / Source method: obtained synthetically |
|---|---|
| #2: Protein | Mass: 13676.723 Da / Num. of mol.: 1 / Fragment: Polybromo Bromdomain 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: PB1, PBRM1 / Production host: Homo sapiens (human) / Gene: PB1, PBRM1 / Production host:   Escherichia coli (E. coli) / Strain (production host): BL21(DE3) / References: UniProt: Q86U86 Escherichia coli (E. coli) / Strain (production host): BL21(DE3) / References: UniProt: Q86U86 |
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||||||||||||||
| Sample conditions | Ionic strength: 0.1 / pH: 6.5 / Pressure: ambient / Temperature: 298 K |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method:  simulated annealing, torsion angle dynamics / Software ordinal: 1 simulated annealing, torsion angle dynamics / Software ordinal: 1 | ||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 200 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller










 PDBj
PDBj




