[English] 日本語
 Yorodumi
Yorodumi- PDB-2k6u: The Solution Structure of a Conformationally Restricted Fully Act... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2k6u | ||||||
|---|---|---|---|---|---|---|---|
| Title | The Solution Structure of a Conformationally Restricted Fully Active Derivative of the Human Relaxin-like Factor (RLF) | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  HORMONE / HORMONE /  protein / protein /  cross-link / cross-link /  isopeptide / Cleavage on pair of basic residues / Disease mutation / Polymorphism / isopeptide / Cleavage on pair of basic residues / Disease mutation / Polymorphism /  Secreted Secreted | ||||||
| Function / homology |  Function and homology information Function and homology informationRelaxin receptors /  oocyte maturation / positive regulation of wound healing / positive regulation of epithelial cell migration / positive regulation of cAMP-mediated signaling / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / G protein-coupled receptor binding / oocyte maturation / positive regulation of wound healing / positive regulation of epithelial cell migration / positive regulation of cAMP-mediated signaling / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / G protein-coupled receptor binding /  insulin receptor binding / insulin receptor binding /  hormone activity / male gonad development ...Relaxin receptors / hormone activity / male gonad development ...Relaxin receptors /  oocyte maturation / positive regulation of wound healing / positive regulation of epithelial cell migration / positive regulation of cAMP-mediated signaling / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / G protein-coupled receptor binding / oocyte maturation / positive regulation of wound healing / positive regulation of epithelial cell migration / positive regulation of cAMP-mediated signaling / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / G protein-coupled receptor binding /  insulin receptor binding / insulin receptor binding /  hormone activity / male gonad development / cell-cell signaling / G alpha (s) signalling events / hormone activity / male gonad development / cell-cell signaling / G alpha (s) signalling events /  spermatogenesis / spermatogenesis /  protease binding / negative regulation of cell population proliferation / protease binding / negative regulation of cell population proliferation /  signaling receptor binding / positive regulation of cell population proliferation / negative regulation of apoptotic process / perinuclear region of cytoplasm / signaling receptor binding / positive regulation of cell population proliferation / negative regulation of apoptotic process / perinuclear region of cytoplasm /  extracellular space / extracellular region extracellular space / extracellular regionSimilarity search - Function | ||||||
| Method |  SOLUTION NMR / torsion angle dynamics, SOLUTION NMR / torsion angle dynamics,  simulated annealing simulated annealing | ||||||
 Authors Authors | Bullesbach, E.E. / Hass, M.A.S. / Jensen, M.R. / Hansen, D.F. / Kristensen, S.M. / Schwabe, C. / Led, J.J. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 2008 Journal: Biochemistry / Year: 2008Title: Solution structure of a conformationally restricted fully active derivative of the human relaxin-like factor Authors: Bullesbach, E.E. / Hass, M.A.S. / Jensen, M.R. / Hansen, D.F. / Kristensen, S.M. / Schwabe, C. / Led, J.J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2k6u.cif.gz 2k6u.cif.gz | 317.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2k6u.ent.gz pdb2k6u.ent.gz | 273.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2k6u.json.gz 2k6u.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/k6/2k6u https://data.pdbj.org/pub/pdb/validation_reports/k6/2k6u ftp://data.pdbj.org/pub/pdb/validation_reports/k6/2k6u ftp://data.pdbj.org/pub/pdb/validation_reports/k6/2k6u | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 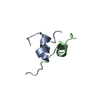
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 2835.240 Da / Num. of mol.: 1 / Fragment: UNP residues 106-131 / Source method: obtained synthetically / Details: This sequence occurs naturally in humans. / References: UniProt: P51460 |
|---|---|
| #2: Protein/peptide | Mass: 3111.731 Da / Num. of mol.: 1 / Fragment: UNP residues 25-51 / Mutation: R50K / Source method: obtained synthetically / Details: This sequence occurs naturally in humans. / References: UniProt: P51460 |
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| |||||||||||||||
| Sample conditions |
|
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: torsion angle dynamics,  simulated annealing / Software ordinal: 1 simulated annealing / Software ordinal: 1 | ||||||||||||||||||||||||||||||||||||||||
| NMR constraints | NOE constraints total: 1050 / NOE intraresidue total count: 457 / NOE long range total count: 182 / NOE medium range total count: 188 / NOE sequential total count: 223 | ||||||||||||||||||||||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 200 / Conformers submitted total number: 20 / Maximum lower distance constraint violation: 0 Å / Maximum upper distance constraint violation: 6 Å / Representative conformer: 1 | ||||||||||||||||||||||||||||||||||||||||
| NMR ensemble rms | Distance rms dev: 0.048 Å / Distance rms dev error: 0.0075 Å |
 Movie
Movie Controller
Controller












 PDBj
PDBj




