[English] 日本語
 Yorodumi
Yorodumi- EMDB-42280: Structure of paused transcription complex Pol II-DSIF-NELF - pois... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of paused transcription complex Pol II-DSIF-NELF - poised post-translocated | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Nucleic acids / Nucleic acids /  transcription / transcription /  RNA polymerase II / NELF / RNA polymerase II / NELF /  DSIF / pausing / TRANSCRIPTION-DNA-RNA complex DSIF / pausing / TRANSCRIPTION-DNA-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationNELF complex / positive regulation of protein modification process / NTRK3 as a dependence receptor / negative regulation of DNA-templated transcription, elongation /  DSIF complex / regulation of transcription elongation by RNA polymerase II / nuclear DNA-directed RNA polymerase complex / B-WICH complex positively regulates rRNA expression / RNA Polymerase I Transcription Initiation / RNA Polymerase I Promoter Escape ...NELF complex / positive regulation of protein modification process / NTRK3 as a dependence receptor / negative regulation of DNA-templated transcription, elongation / DSIF complex / regulation of transcription elongation by RNA polymerase II / nuclear DNA-directed RNA polymerase complex / B-WICH complex positively regulates rRNA expression / RNA Polymerase I Transcription Initiation / RNA Polymerase I Promoter Escape ...NELF complex / positive regulation of protein modification process / NTRK3 as a dependence receptor / negative regulation of DNA-templated transcription, elongation /  DSIF complex / regulation of transcription elongation by RNA polymerase II / nuclear DNA-directed RNA polymerase complex / B-WICH complex positively regulates rRNA expression / RNA Polymerase I Transcription Initiation / RNA Polymerase I Promoter Escape / RNA Polymerase I Transcription Termination / RNA Polymerase III Transcription Initiation From Type 1 Promoter / RNA Polymerase III Transcription Initiation From Type 2 Promoter / RNA Polymerase III Transcription Initiation From Type 3 Promoter / Formation of RNA Pol II elongation complex / Formation of the Early Elongation Complex / Transcriptional regulation by small RNAs / RNA Polymerase II Pre-transcription Events / TP53 Regulates Transcription of DNA Repair Genes / FGFR2 alternative splicing / RNA polymerase II transcribes snRNA genes / mRNA Capping / mRNA Splicing - Major Pathway / mRNA Splicing - Minor Pathway / Processing of Capped Intron-Containing Pre-mRNA / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Elongation / RNA Polymerase II Transcription Initiation And Promoter Clearance / RNA Pol II CTD phosphorylation and interaction with CE / Estrogen-dependent gene expression / Formation of TC-NER Pre-Incision Complex / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER / negative regulation of stem cell differentiation / nuclear lumen / Abortive elongation of HIV-1 transcript in the absence of Tat / positive regulation of DNA-templated transcription, elongation / transcription elongation-coupled chromatin remodeling / DSIF complex / regulation of transcription elongation by RNA polymerase II / nuclear DNA-directed RNA polymerase complex / B-WICH complex positively regulates rRNA expression / RNA Polymerase I Transcription Initiation / RNA Polymerase I Promoter Escape / RNA Polymerase I Transcription Termination / RNA Polymerase III Transcription Initiation From Type 1 Promoter / RNA Polymerase III Transcription Initiation From Type 2 Promoter / RNA Polymerase III Transcription Initiation From Type 3 Promoter / Formation of RNA Pol II elongation complex / Formation of the Early Elongation Complex / Transcriptional regulation by small RNAs / RNA Polymerase II Pre-transcription Events / TP53 Regulates Transcription of DNA Repair Genes / FGFR2 alternative splicing / RNA polymerase II transcribes snRNA genes / mRNA Capping / mRNA Splicing - Major Pathway / mRNA Splicing - Minor Pathway / Processing of Capped Intron-Containing Pre-mRNA / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Elongation / RNA Polymerase II Transcription Initiation And Promoter Clearance / RNA Pol II CTD phosphorylation and interaction with CE / Estrogen-dependent gene expression / Formation of TC-NER Pre-Incision Complex / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER / negative regulation of stem cell differentiation / nuclear lumen / Abortive elongation of HIV-1 transcript in the absence of Tat / positive regulation of DNA-templated transcription, elongation / transcription elongation-coupled chromatin remodeling /  RNA polymerase complex / RNA Pol II CTD phosphorylation and interaction with CE during HIV infection / RNA Pol II CTD phosphorylation and interaction with CE / Formation of the Early Elongation Complex / Formation of the HIV-1 Early Elongation Complex / mRNA Capping / maintenance of transcriptional fidelity during transcription elongation by RNA polymerase II / negative regulation of transcription elongation by RNA polymerase II / Pausing and recovery of Tat-mediated HIV elongation / Tat-mediated HIV elongation arrest and recovery / RNA polymerase complex / RNA Pol II CTD phosphorylation and interaction with CE during HIV infection / RNA Pol II CTD phosphorylation and interaction with CE / Formation of the Early Elongation Complex / Formation of the HIV-1 Early Elongation Complex / mRNA Capping / maintenance of transcriptional fidelity during transcription elongation by RNA polymerase II / negative regulation of transcription elongation by RNA polymerase II / Pausing and recovery of Tat-mediated HIV elongation / Tat-mediated HIV elongation arrest and recovery /  RNA polymerase II activity / positive regulation of macroautophagy / organelle membrane / RNA polymerase II transcribes snRNA genes / HIV elongation arrest and recovery / Pausing and recovery of HIV elongation / transcription-coupled nucleotide-excision repair / Tat-mediated elongation of the HIV-1 transcript / Formation of HIV-1 elongation complex containing HIV-1 Tat / RNA polymerase II activity / positive regulation of macroautophagy / organelle membrane / RNA polymerase II transcribes snRNA genes / HIV elongation arrest and recovery / Pausing and recovery of HIV elongation / transcription-coupled nucleotide-excision repair / Tat-mediated elongation of the HIV-1 transcript / Formation of HIV-1 elongation complex containing HIV-1 Tat /  RNA polymerase I complex / RNA polymerase I complex /  RNA polymerase III complex / Formation of HIV elongation complex in the absence of HIV Tat / localization / RNA polymerase III complex / Formation of HIV elongation complex in the absence of HIV Tat / localization /  RNA polymerase II, core complex / RNA Polymerase II Transcription Elongation / Formation of RNA Pol II elongation complex / RNA Polymerase II Pre-transcription Events / RNA polymerase II, core complex / RNA Polymerase II Transcription Elongation / Formation of RNA Pol II elongation complex / RNA Polymerase II Pre-transcription Events /  DNA-directed RNA polymerase complex / transcription elongation by RNA polymerase II / DNA-directed RNA polymerase complex / transcription elongation by RNA polymerase II /  stem cell differentiation / stem cell differentiation /  transcription initiation at RNA polymerase II promoter / DNA-templated transcription initiation / TP53 Regulates Transcription of DNA Repair Genes / transcription initiation at RNA polymerase II promoter / DNA-templated transcription initiation / TP53 Regulates Transcription of DNA Repair Genes /  ribonucleoside binding / ribonucleoside binding /  fibrillar center / DNA-directed 5'-3' RNA polymerase activity / fibrillar center / DNA-directed 5'-3' RNA polymerase activity /  DNA-directed RNA polymerase / cell population proliferation / transcription by RNA polymerase II / DNA-directed RNA polymerase / cell population proliferation / transcription by RNA polymerase II /  nucleic acid binding / positive regulation of ERK1 and ERK2 cascade / molecular adaptor activity / nucleic acid binding / positive regulation of ERK1 and ERK2 cascade / molecular adaptor activity /  nuclear body / nuclear body /  protein dimerization activity / protein heterodimerization activity / protein dimerization activity / protein heterodimerization activity /  nucleotide binding / nucleotide binding /  mRNA binding / DNA-templated transcription / mRNA binding / DNA-templated transcription /  chromatin binding / chromatin binding /  chromatin / chromatin /  nucleolus / negative regulation of transcription by RNA polymerase II / nucleolus / negative regulation of transcription by RNA polymerase II /  enzyme binding / positive regulation of transcription by RNA polymerase II / enzyme binding / positive regulation of transcription by RNA polymerase II /  DNA binding / DNA binding /  RNA binding / zinc ion binding / RNA binding / zinc ion binding /  nucleoplasm / nucleoplasm /  membrane / membrane /  metal ion binding metal ion bindingSimilarity search - Function | |||||||||
| Biological species |   Sus scrofa (pig) / Sus scrofa (pig) /   Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.7 Å cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Vos SM / Su BG | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Distinct negative elongation factor conformations regulate RNA polymerase II promoter-proximal pausing. Authors: Bonnie G Su / Seychelle M Vos /  Abstract: Metazoan gene expression regulation involves pausing of RNA polymerase (Pol II) in the promoter-proximal region of genes and is stabilized by DSIF and NELF. Upon depletion of elongation factors, NELF ...Metazoan gene expression regulation involves pausing of RNA polymerase (Pol II) in the promoter-proximal region of genes and is stabilized by DSIF and NELF. Upon depletion of elongation factors, NELF appears to accompany elongating Pol II past pause sites; however, prior work indicates that NELF prevents Pol II elongation. Here, we report cryoelectron microscopy structures of Pol II-DSIF-NELF complexes with NELF in two distinct conformations corresponding to paused and poised states. The paused NELF state supports Pol II stalling, whereas the poised NELF state enables transcription elongation as it does not support a tilted RNA-DNA hybrid. Further, the poised NELF state can accommodate TFIIS binding to Pol II, allowing for Pol II reactivation at paused or backtracking sites. Finally, we observe that the NELF-A tentacle interacts with the RPB2 protrusion and is necessary for pausing. Our results define how NELF can support pausing, reactivation, and elongation by Pol II. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42280.map.gz emd_42280.map.gz | 260.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42280-v30.xml emd-42280-v30.xml emd-42280.xml emd-42280.xml | 40 KB 40 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_42280.png emd_42280.png | 148.9 KB | ||
| Masks |  emd_42280_msk_1.map emd_42280_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-42280.cif.gz emd-42280.cif.gz | 11.7 KB | ||
| Others |  emd_42280_additional_1.map.gz emd_42280_additional_1.map.gz emd_42280_half_map_1.map.gz emd_42280_half_map_1.map.gz emd_42280_half_map_2.map.gz emd_42280_half_map_2.map.gz | 242.6 MB 475.4 MB 475.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42280 http://ftp.pdbj.org/pub/emdb/structures/EMD-42280 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42280 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42280 | HTTPS FTP |
-Related structure data
| Related structure data |  8uhgMC  8uhaC  8uhdC  8ui0C  8uisC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42280.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42280.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.822 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_42280_msk_1.map emd_42280_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
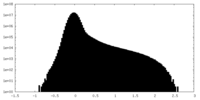
| Density Histograms |
-Additional map: #1
| File | emd_42280_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
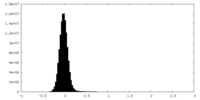
| Density Histograms |
-Half map: #1
| File | emd_42280_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
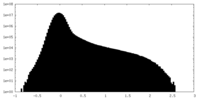
| Density Histograms |
-Half map: #2
| File | emd_42280_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : RNA Polymerase in complex with DSIF and NELF
+Supramolecule #1: RNA Polymerase in complex with DSIF and NELF
+Macromolecule #1: Negative elongation factor E
+Macromolecule #2: Negative elongation factor B
+Macromolecule #3: Negative elongation factor C/D
+Macromolecule #4: DNA-directed RNA polymerase subunit
+Macromolecule #5: DNA-directed RNA polymerase subunit beta
+Macromolecule #6: DNA-directed RNA polymerase II subunit RPB3
+Macromolecule #7: RNA polymerase Rpb4/RPC9 core domain-containing protein
+Macromolecule #8: DNA-directed RNA polymerase II subunit E
+Macromolecule #9: DNA-directed RNA polymerases I, II, and III subunit RPABC2
+Macromolecule #10: DNA-directed RNA polymerase subunit
+Macromolecule #11: DNA-directed RNA polymerases I, II, and III subunit RPABC3
+Macromolecule #12: DNA-directed RNA polymerase II subunit RPB9
+Macromolecule #13: DNA-directed RNA polymerases I, II, and III subunit RPABC5
+Macromolecule #14: DNA-directed RNA polymerase II subunit RPB11-a
+Macromolecule #15: RNA polymerase II subunit K
+Macromolecule #19: Negative elongation factor A
+Macromolecule #20: Transcription elongation factor SPT5
+Macromolecule #16: Non-template DNA
+Macromolecule #18: Template DNA
+Macromolecule #17: RNA
+Macromolecule #21: ZINC ION
+Macromolecule #22: MAGNESIUM ION
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 51.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.7 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 90283 |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)