+ Open data
Open data
- Basic information
Basic information
| Entry | 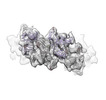 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Sub-tomogram average of pro-meprin alpha supercoiled filament | |||||||||
 Map data Map data | Full reconstruction (unsharpened, masked, filtered to global 0.143 FSC) | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | subtomogram averaging /  cryo EM / Resolution: 12.7 Å cryo EM / Resolution: 12.7 Å | |||||||||
 Authors Authors | Bayly-Jones C / Lupton CJ / Fritz C / Schlenzig D / Whisstock JC | |||||||||
| Funding support |  Germany, Germany,  Australia, 2 items Australia, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Helical ultrastructure of the metalloprotease meprin α in complex with a small molecule inhibitor. Authors: Charles Bayly-Jones / Christopher J Lupton / Claudia Fritz / Hariprasad Venugopal / Daniel Ramsbeck / Michael Wermann / Christian Jäger / Alex de Marco / Stephan Schilling / Dagmar ...Authors: Charles Bayly-Jones / Christopher J Lupton / Claudia Fritz / Hariprasad Venugopal / Daniel Ramsbeck / Michael Wermann / Christian Jäger / Alex de Marco / Stephan Schilling / Dagmar Schlenzig / James C Whisstock /   Abstract: The zinc-dependent metalloprotease meprin α is predominantly expressed in the brush border membrane of proximal tubules in the kidney and enterocytes in the small intestine and colon. In normal ...The zinc-dependent metalloprotease meprin α is predominantly expressed in the brush border membrane of proximal tubules in the kidney and enterocytes in the small intestine and colon. In normal tissue homeostasis meprin α performs key roles in inflammation, immunity, and extracellular matrix remodelling. Dysregulated meprin α is associated with acute kidney injury, sepsis, urinary tract infection, metastatic colorectal carcinoma, and inflammatory bowel disease. Accordingly, meprin α is the target of drug discovery programs. In contrast to meprin β, meprin α is secreted into the extracellular space, whereupon it oligomerises to form giant assemblies and is the largest extracellular protease identified to date (~6 MDa). Here, using cryo-electron microscopy, we determine the high-resolution structure of the zymogen and mature form of meprin α, as well as the structure of the active form in complex with a prototype small molecule inhibitor and human fetuin-B. Our data reveal that meprin α forms a giant, flexible, left-handed helical assembly of roughly 22 nm in diameter. We find that oligomerisation improves proteolytic and thermal stability but does not impact substrate specificity or enzymatic activity. Furthermore, structural comparison with meprin β reveal unique features of the active site of meprin α, and helical assembly more broadly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27689.map.gz emd_27689.map.gz | 2.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27689-v30.xml emd-27689-v30.xml emd-27689.xml emd-27689.xml | 23.3 KB 23.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27689_fsc.xml emd_27689_fsc.xml | 3.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_27689.png emd_27689.png | 76.9 KB | ||
| Masks |  emd_27689_msk_1.map emd_27689_msk_1.map | 3.4 MB |  Mask map Mask map | |
| Others |  emd_27689_additional_1.map.gz emd_27689_additional_1.map.gz emd_27689_additional_2.map.gz emd_27689_additional_2.map.gz emd_27689_half_map_1.map.gz emd_27689_half_map_1.map.gz emd_27689_half_map_2.map.gz emd_27689_half_map_2.map.gz | 3.1 MB 741.9 KB 2.4 MB 2.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27689 http://ftp.pdbj.org/pub/emdb/structures/EMD-27689 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27689 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27689 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27689.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27689.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full reconstruction (unsharpened, masked, filtered to global 0.143 FSC) | ||||||||||||||||||||
| Voxel size | X=Y=Z: 4.5 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27689_msk_1.map emd_27689_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: DeepEMhancer sharpened map (wide mode)
| File | emd_27689_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer sharpened map (wide mode) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: B-factor sharpened map (Relion)
| File | emd_27689_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | B-factor sharpened map (Relion) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1 of 2
| File | emd_27689_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 of 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2 of 2
| File | emd_27689_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 of 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Roughly 12 subunits of helical meprin alpha in the zymogen state
| Entire | Name: Roughly 12 subunits of helical meprin alpha in the zymogen state |
|---|---|
| Components |
|
-Supramolecule #1: Roughly 12 subunits of helical meprin alpha in the zymogen state
| Supramolecule | Name: Roughly 12 subunits of helical meprin alpha in the zymogen state type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: all Details: Sub-tomogram average of a ~12 subunit region of recombinant, secreted helical pro-meprin alpha |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Drosophila (fruit flies) / Recombinant cell: Schneider-2 / Recombinant plasmid: pMT/BiP/V5 Drosophila (fruit flies) / Recombinant cell: Schneider-2 / Recombinant plasmid: pMT/BiP/V5 |
| Molecular weight | Theoretical: 85 kDa/nm |
-Macromolecule #1: Pro-meprin alpha
| Macromolecule | Name: Pro-meprin alpha / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Drosophila melanogaster (fruit fly) Drosophila melanogaster (fruit fly) |
| Sequence | String: WSHPQFEKVP IKYLPEENVH DADFGEQKDI SEINLAAGLD LFQGDIL LQ KSRNGLRDPN TRWTFPIPYI LADNLGLNAK GAILYAFEMF RLKSCVDFKP YEGESSYI I FQQFDGCWSE VGDQHVGQNI SIGQGCAYKA IIEHEILHAL GFYHEQSRTD RDDYVNIWW ...String: WSHPQFEKVP IKYLPEENVH DADFGEQKDI SEINLAAGLD LFQGDIL LQ KSRNGLRDPN TRWTFPIPYI LADNLGLNAK GAILYAFEMF RLKSCVDFKP YEGESSYI I FQQFDGCWSE VGDQHVGQNI SIGQGCAYKA IIEHEILHAL GFYHEQSRTD RDDYVNIWW DQILSGYQHN FDTYDDSLIT DLNTPYDYES LMHYQPFSFN KNASVPTITA KIPEFNSIIG QRLDFSAID LERLNRMYNC TTTHTLLDHC TFEKANICGM IQGTRDDTDW AHQDSAQAGE V DHTLLGQC TGAGYFMQFS TSSGSAEEAA LLESRILYPK RKQQCLQFFY KMTGSPSDRL VV WVRRDDS TGNVRKLVKV QTFQGDDDHN WKIAHVVLKE EQKFRYLFQG TKGDPQNSTG GIY LDDITL TETPCPTGVW TVRNFSQVLE NTSKGDKLQS PRFYNSEGYG FGVTLYPNSR ESSG YLRLA FHVCSGENDA ILEWPVENRQ VIITILDQEP DVRNRMSSSM VFTTSKSHTS PAIND TVIW DRPSRVGTYH TDCNCFRSID LGWSGFISHQ MLKRRSFLKN DDLIIFVDFE DITHLS QTE VPTKGKRLSP QGLILQGQEQ QVSEEGSGKA MLEEALPVSL SQGQPSRQKR SVENTGP LE DHNWPQYFRD PCDPNPCQND GICVNVKGMA SCRCISGHAF FYTGERCQAV QVHGSVLG M VIGGTAGVIF LTFSIIAILS QRPRK |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV / Details: 3 s blot, -3 force. | |||||||||
| Details | Polydisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.2 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 64000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.2 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 64000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Average electron dose: 2.5 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z
Z Y
Y X
X










































