+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
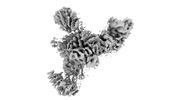
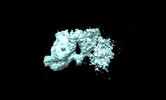
| Title | Mediator-PIC Early (Tail A) | |||||||||
 Map data Map data | Med-PIC Tail A - Unsharpened | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationTFIIE-class transcription factor complex binding / regulation of establishment of protein localization to chromosome / positive regulation of invasive growth in response to glucose limitation / TFIIH-class transcription factor complex binding / core mediator complex /  mediator complex / TFIIB-class transcription factor binding / positive regulation of transcription initiation by RNA polymerase II / RNA polymerase II core promoter sequence-specific DNA binding / RNA polymerase II preinitiation complex assembly ...TFIIE-class transcription factor complex binding / regulation of establishment of protein localization to chromosome / positive regulation of invasive growth in response to glucose limitation / TFIIH-class transcription factor complex binding / core mediator complex / mediator complex / TFIIB-class transcription factor binding / positive regulation of transcription initiation by RNA polymerase II / RNA polymerase II core promoter sequence-specific DNA binding / RNA polymerase II preinitiation complex assembly ...TFIIE-class transcription factor complex binding / regulation of establishment of protein localization to chromosome / positive regulation of invasive growth in response to glucose limitation / TFIIH-class transcription factor complex binding / core mediator complex /  mediator complex / TFIIB-class transcription factor binding / positive regulation of transcription initiation by RNA polymerase II / RNA polymerase II core promoter sequence-specific DNA binding / RNA polymerase II preinitiation complex assembly / positive regulation of transcription elongation by RNA polymerase II / mediator complex / TFIIB-class transcription factor binding / positive regulation of transcription initiation by RNA polymerase II / RNA polymerase II core promoter sequence-specific DNA binding / RNA polymerase II preinitiation complex assembly / positive regulation of transcription elongation by RNA polymerase II /  transcription coregulator activity / RNA polymerase II-specific DNA-binding transcription factor binding / transcription by RNA polymerase II / transcription coregulator activity / RNA polymerase II-specific DNA-binding transcription factor binding / transcription by RNA polymerase II /  transcription coactivator activity / regulation of transcription by RNA polymerase II / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / transcription coactivator activity / regulation of transcription by RNA polymerase II / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II /  mitochondrion / mitochondrion /  nucleus / nucleus /  cytosol cytosolSimilarity search - Function | |||||||||
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) / Saccharomyces cerevisiae (brewer's yeast) /   Saccharomyces cerevisiae S288C (yeast) Saccharomyces cerevisiae S288C (yeast) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.7 Å cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Gorbea Colon JJ / Chen S-F / Tsai KL / Murakami K | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2023 Journal: Mol Cell / Year: 2023Title: Structural basis of a transcription pre-initiation complex on a divergent promoter. Authors: Jose J Gorbea Colón / Leon Palao / Shin-Fu Chen / Hee Jong Kim / Laura Snyder / Yi-Wei Chang / Kuang-Lei Tsai / Kenji Murakami /  Abstract: Most eukaryotic promoter regions are divergently transcribed. As the RNA polymerase II pre-initiation complex (PIC) is intrinsically asymmetric and responsible for transcription in a single ...Most eukaryotic promoter regions are divergently transcribed. As the RNA polymerase II pre-initiation complex (PIC) is intrinsically asymmetric and responsible for transcription in a single direction, it is unknown how divergent transcription arises. Here, the Saccharomyces cerevisiae Mediator complexed with a PIC (Med-PIC) was assembled on a divergent promoter and analyzed by cryoelectron microscopy. The structure reveals two distinct Med-PICs forming a dimer through the Mediator tail module, induced by a homodimeric activator protein localized near the dimerization interface. The tail dimer is associated with ∼80-bp upstream DNA, such that two flanking core promoter regions are positioned and oriented in a suitable form for PIC assembly in opposite directions. Also, cryoelectron tomography visualized the progress of the PIC assembly on the two core promoter regions, providing direct evidence for the role of the Med-PIC dimer in divergent transcription. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26543.map.gz emd_26543.map.gz | 65 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26543-v30.xml emd-26543-v30.xml emd-26543.xml emd-26543.xml | 28.5 KB 28.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26543_fsc.xml emd_26543_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_26543.png emd_26543.png | 38.6 KB | ||
| Masks |  emd_26543_msk_1.map emd_26543_msk_1.map | 83.7 MB |  Mask map Mask map | |
| Others |  emd_26543_additional_1.map.gz emd_26543_additional_1.map.gz emd_26543_additional_2.map.gz emd_26543_additional_2.map.gz emd_26543_half_map_1.map.gz emd_26543_half_map_1.map.gz emd_26543_half_map_2.map.gz emd_26543_half_map_2.map.gz | 76.5 MB 3.1 MB 65.2 MB 65.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26543 http://ftp.pdbj.org/pub/emdb/structures/EMD-26543 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26543 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26543 | HTTPS FTP |
-Related structure data
| Related structure data |  7uicMC  7ui9C  7uifC  7uigC  7uikC  7uilC  7uioC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26543.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26543.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Med-PIC Tail A - Unsharpened | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.38 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26543_msk_1.map emd_26543_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Med-PIC Tail A - Sharpened (Relion)
| File | emd_26543_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Med-PIC Tail A - Sharpened (Relion) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Med-PIC Tail A - Sharpened (DeepEMhancer)
| File | emd_26543_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Med-PIC Tail A - Sharpened (DeepEMhancer) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Med-PIC Tail A - Half 2
| File | emd_26543_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Med-PIC Tail A - Half 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Med-PIC Tail A - Half 1
| File | emd_26543_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Med-PIC Tail A - Half 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Mediator-PIC Early
| Entire | Name: Mediator-PIC Early |
|---|---|
| Components |
|
-Supramolecule #1: Mediator-PIC Early
| Supramolecule | Name: Mediator-PIC Early / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) |
-Macromolecule #1: Mediator of RNA polymerase II transcription subunit 2
| Macromolecule | Name: Mediator of RNA polymerase II transcription subunit 2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae S288C (yeast) / Strain: ATCC 204508 / S288c Saccharomyces cerevisiae S288C (yeast) / Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 47.75427 KDa |
| Sequence | String: MVVQNSPVSS VHTANFSERG SNTRTMTYKN KLTVCFDDIL KVGAEMMMQQ QLKNVQLDSY LVNGFSQSQQ KLLKEKVKLF HGILDDLET SLSQSSSYLE TLTALGKEKE KEREEAEKKR AEQENMRKVR EQEELKKRQE LEEASQQQQL QQNSKEKNGL G LNFSTTAP ...String: MVVQNSPVSS VHTANFSERG SNTRTMTYKN KLTVCFDDIL KVGAEMMMQQ QLKNVQLDSY LVNGFSQSQQ KLLKEKVKLF HGILDDLET SLSQSSSYLE TLTALGKEKE KEREEAEKKR AEQENMRKVR EQEELKKRQE LEEASQQQQL QQNSKEKNGL G LNFSTTAP ANTTDANGSK ENYQELGSLQ SSSQTQLENA NAANNGAAFS PLTTTRIQSQ QAQPSDVMFN DLNSMDISMF SG LDSTGFD STAFNATVDE TKGFDDNDSG NNYNDINISS IENNINNNIN STKNGKDNNN ESNKNNNGDE KNKNNNEDNE NNN NSSEKN NNNNNNNNNN NDDNGNNNNN NSGNDNNNTT NNDSNNKNNS ITTGNDNENI VNNDLPTTVV SNPGDNPPPA DNGE EYLTL NDFNDLNIDW STTGDNGELD LSGFNI |
-Macromolecule #2: Mediator of RNA polymerase II transcription subunit 3
| Macromolecule | Name: Mediator of RNA polymerase II transcription subunit 3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae S288C (yeast) / Strain: ATCC 204508 / S288c Saccharomyces cerevisiae S288C (yeast) / Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 43.116039 KDa |
| Sequence | String: MDSIIPAGVK LDDLQVILAK NENETRDKVC KQINEARDEI LPLRLQFNEF IQIMANIDQE GSKQADRMAK YLHIRDKILQ LNDRFQTLS SHLEALQPLF STVPEYLKTA DNRDRSFQLL EPLSTYNKNG NAVCSTATVV STNHSAAAST PTTTATPHAN P ITHAHSLS ...String: MDSIIPAGVK LDDLQVILAK NENETRDKVC KQINEARDEI LPLRLQFNEF IQIMANIDQE GSKQADRMAK YLHIRDKILQ LNDRFQTLS SHLEALQPLF STVPEYLKTA DNRDRSFQLL EPLSTYNKNG NAVCSTATVV STNHSAAAST PTTTATPHAN P ITHAHSLS NPNSTATMQH NPLAGKRGPK SGSTMGTPTV HNSTAAAPIA APKKPRKPRQ TKKAKAQAQA QAQAQAQVYA QQ STVQTPI TASMAAALPN PTPSMINSVS PTNVMGTPLT NMMSPMGNAY SMGAQNQGGQ VSMSQFNGSG NGSNPNTNTN SNN TPLQSQ LNLNNLTPAN ILNMSMNNDF QQQQQQQQQQ QQPQPQYNMN MGMNNMNNGG KELDSLDLNN LELGGLNMDF L |
-Macromolecule #3: Mediator of RNA polymerase II transcription subunit 5
| Macromolecule | Name: Mediator of RNA polymerase II transcription subunit 5 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae S288C (yeast) / Strain: ATCC 204508 / S288c Saccharomyces cerevisiae S288C (yeast) / Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 128.91875 KDa |
| Sequence | String: MEKESVYNLA LKCAERQLTS MEFSNLYKEF FNEKFPSLIQ EEEEDTTTTA NINEVKKASD LVDTPSNNTA ATADTTHLHE ALDIVCSDF VKILNLEKPL ILADYIVEVL LVNYNSDMIK CFLPKLNSVR NSLLLAHFFS KSCSFFAKLS DTLIIDQVRK D LGNVIVPN ...String: MEKESVYNLA LKCAERQLTS MEFSNLYKEF FNEKFPSLIQ EEEEDTTTTA NINEVKKASD LVDTPSNNTA ATADTTHLHE ALDIVCSDF VKILNLEKPL ILADYIVEVL LVNYNSDMIK CFLPKLNSVR NSLLLAHFFS KSCSFFAKLS DTLIIDQVRK D LGNVIVPN ILSLDMNSMN KELIAIVSKL LQTTLKLSPS PILLTSAGCK NGSFTLLNQL SQTNKLLFKR VSQTFEAKLH FK DTKPFLN KDSTNEFVGS PSLTSPQYIP SPLSSTKPPG SVNSAAKYKD MKLLRYYKNI WLNNKIINWE ISNPDFLSKY SAI TSSIFQ ESFNSVQNLD QLLTDLIETS FTCFAQFVSN KQYHQANSNL TLLERKWVIF ITKHLPLLIL ENSSRSPRVV TNAL DNIDE KVVKAIRIYF TEKDDNKTNN EDLFDDYPST SLDIRHDFIK GLIMLNLQPA SVINNYLRED QMIDTSILPT RDDLF VRNL QGIQEVVHNT NSFIISSLDT LELESITESI THDSSNGLFQ VLHNFESVAP TKQREIVKAF LSIFEDAIKE LNYNRI AKI CALLFFNFSH SLTTILSFSS PAALMKTLIK FVDLSRNGRN GSNGNDESSE YETINISLSF SWAILLIINL TQTYGIS VV DVALKYPELS IKNSFIINFI SNLPNVSDKY YLEESNVNDS DMLTKSHNTV QSWLCDLFVN GSITDQLIQN IETRQLAN L IPFIVKQVLL SVEIGVLTDI SSLIGGFEYF LQPLLLVGLI KTFYWLEQFL SCVKNDTISE DILQGIFNLL NTLFNPVTL NEDSKAFHTA VLRLNAIPLL KVLRKFRVQS QSNYGIYSSD AQGDPNLEPL IAKLVAVLNV SPVYDVDPRI INSENDYSRK QLGYGKFLI LNENPINKIM TNQINSFWSL HSSTYYNLDY LFELIELVTP KSFLFDVLKT LEYKLATYGV PGSENKRGSL D SEHVFDYF FYFLVLYDVK TAEEASQLIE YMENDAKKSK GDVDIKGEDL HEKNDSAEVR QETQPKAEAT QDDDFDMLFG EN DTSTQAY EEEEENEDND GNNRTNNVPM IKAEETPSKT NKISILKRHS FAVLLHERKL LNDLALENGE ITKTENEKFI SYH DKYLCM LKTCVF |
-Macromolecule #4: Mediator of RNA polymerase II transcription subunit 14
| Macromolecule | Name: Mediator of RNA polymerase II transcription subunit 14 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae S288C (yeast) / Strain: ATCC 204508 / S288c Saccharomyces cerevisiae S288C (yeast) / Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 123.500695 KDa |
| Sequence | String: MTTTIGSPQM LANEERLSNE MHALKNRSEQ NGQEQQGPVK NTQLHGPSAT DPETTATQKE SLEMVPKDTS AATMTSAPPP ALPHVEINQ VSLALVIRNL TVFTMKELAQ YMKTNVHTQA NEPNSAKKIR FLQLIIFLRT QFLKLYVLVK WTRTIKQNNF H VLIDLLNW ...String: MTTTIGSPQM LANEERLSNE MHALKNRSEQ NGQEQQGPVK NTQLHGPSAT DPETTATQKE SLEMVPKDTS AATMTSAPPP ALPHVEINQ VSLALVIRNL TVFTMKELAQ YMKTNVHTQA NEPNSAKKIR FLQLIIFLRT QFLKLYVLVK WTRTIKQNNF H VLIDLLNW FRTTNMNVNN CIWALKSSLN SMTNAKLPNV DLVTALEVLS LGRPNLPTHN FKLSGVSNSM DMVDGMAKVP IG LILQRLK DLNLTVSIKI ALMNIPKPLN SYHIKNGRIY FTVPNEFEIQ LSTVNRQSPL FFVDLKLLFN TEAEQTVSAV TEA TSTNGD SENNEENSSS NGNNLPLNKP RLEKLINEIL LKSNDPLLSL YNFLHKYVLT LQLYMVHREF LKLANGGKFS KSNL IHNYD SKKSTITVRY WLNGKMDSKG KITIGIQRTT ESLILKWDNQ SASRAKNMPV IYNNIVSNIE GILDEIMFNH ARIIR SELL ARDIFQEDEE NSDVLLFQLP TTCVSMAPIQ LKIDLLSGQF YFRNPTPLLS NYASKINRAE GPEELARILQ QLKLDK IIH VLTTMFENTG WSCSRIIKID KPIRTQVNTG GESVVKKEDN KYAIAGNSTT NSDVSLLLQR DLFIRLPHWP LNWYLIL SI ISSKTSCVVE KRIGKIVSQR GKWNLKYLDN SNVMTVKLES ITYQKIMILQ RTILNRIINH MLIDSLNQLE IRNKICSS E MINEQKLPQY IIQGSNTNDN ISIITLELES FLEGSKALNS ILESSMFLRI DYSNSQIRLY AKFKRNTMMI QCQIDKLYI HFVQEEPLAF YLEESFTNLG IIVQYLTKFR QKLMQLVVLT DVVERLHKNF ESENFKIIAL QPNEISFKYL SNNDEDDKDC TIKISTNDD SIKNLTVQLS PSNPQHIIQP FLDNSKMDYH FIFSYLQFTS SLFKALKVIL NERGGKFHES GSQYSTMVNI G LHNLNEYQ IVYYNPQAGT KITICIELKT VLHNGRDKIQ FHIHFADVAH ITTKSPAYPM MHQVRNQVFM LDTKRLGTPE SV KPANASH AIRLGNGVAC DPSEIEPILM EIHNILKVDS NSSSS |
-Macromolecule #5: Mediator of RNA polymerase II transcription subunit 15
| Macromolecule | Name: Mediator of RNA polymerase II transcription subunit 15 type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae S288C (yeast) / Strain: ATCC 204508 / S288c Saccharomyces cerevisiae S288C (yeast) / Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 120.432242 KDa |
| Sequence | String: MSAAPVQDKD TLSNAERAKN VNGLLQVLMD INTLNGGSSD TADKIRIHAK NFEAALFAKS SSKKEYMDSM NEKVAVMRNT YNTRKNAVT AAAANNNIKP VEQHHINNLK NSGNSANNMN VNMNLNPQMF LNQQAQARQQ VAQQLRNQQQ QQQQQQQQQR R QLTPQQQQ ...String: MSAAPVQDKD TLSNAERAKN VNGLLQVLMD INTLNGGSSD TADKIRIHAK NFEAALFAKS SSKKEYMDSM NEKVAVMRNT YNTRKNAVT AAAANNNIKP VEQHHINNLK NSGNSANNMN VNMNLNPQMF LNQQAQARQQ VAQQLRNQQQ QQQQQQQQQR R QLTPQQQQ LVNQMKVAPI PKQLLQRIPN IPPNINTWQQ VTALAQQKLL TPQDMEAAKE VYKIHQQLLF KARLQQQQAQ AQ AQANNNN NGLPQNGNIN NNINIPQQQQ MQPPNSSANN NPLQQQSSQN TVPNVLNQIN QIFSPEEQRS LLQEAIETCK NFE KTQLGS TMTEPVKQSF IRKYINQKAL RKIQALRDVK NNNNANNNGS NLQRAQNVPM NIIQQQQQQN TNNNDTIATS ATPN AAAFS QQQNASSKLY QMQQQQQAQA QAQAQAQAQA QAQAQAQAAQ AAQAQAQAQA QAQAQAQAQA QAQAQAQAQA QAQAQ AHAQ HQPSQQPQQA QQQPNPLHGL TPTAKDVEVI KQLSLDASKT NLRLTDVTNS LSNEEKEKIK MKLKQGQKLF VQVSNF APQ VYIITKNENF LKEVFQLRIF VKEILEKCAE GIFVVKLDTV DRLIIKYQKY WESMRIQILR RQAILRQQQQ MANNNGN PG TTSTGNNNNI ATQQNMQQSL QQMQHLQQLK MQQQQQQQQQ QQQQQQQQQQ QQQQHIYPSS TPGVANYSAM ANAPGNNI P YMNHKNTSSM DFLNSMENTP KVPVSAAATP SLNKTINGKV NGRTKSNTIP VTSIPSTNKK LSISNAASQQ PTPRSASNT AKSTPNTNPS PLKTQTKNGT PNPNNMKTVQ SPMGAQPSYN SAIIENAFRK EELLLKDLEI RKLEISSRFK HRQEIFKDSP MDLFMSTLG DCLGIKDEEM LTSCTIPKAV VDHINGSGKR KPTKAAQRAR DQDSIDISIK DNKLVMKSKF NKSNRSYSIA L SNVAAIFK GIGGNFKDLS TLVHSSSPST SSNMDVGNPR KRKASVLEIS PQDSIASVLS PDSNIMSDSK KIKVDSPDDP FM TKSGATT SEKQEVTNEA PFLTSGTSSE QFNVWDWNNW TSAT |
-Macromolecule #6: Mediator of RNA polymerase II transcription subunit 16
| Macromolecule | Name: Mediator of RNA polymerase II transcription subunit 16 type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae S288C (yeast) / Strain: ATCC 204508 / S288c Saccharomyces cerevisiae S288C (yeast) / Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 111.423422 KDa |
| Sequence | String: MMLGEHLMSW SKTGIIAYSD SQSSNANICL TFLESINGIN WRFHTPQKYV LHPQLHEVQY QESSSTLSTH STTTSVNGST TAGVGSTPN FGGNSNKSPP QFFYNISSIH WNNWFSLPGD MLAVCDELGN MTMLITGQRP DRATTYEKLT MVFQDNVYKI Y NHVMPLKP ...String: MMLGEHLMSW SKTGIIAYSD SQSSNANICL TFLESINGIN WRFHTPQKYV LHPQLHEVQY QESSSTLSTH STTTSVNGST TAGVGSTPN FGGNSNKSPP QFFYNISSIH WNNWFSLPGD MLAVCDELGN MTMLITGQRP DRATTYEKLT MVFQDNVYKI Y NHVMPLKP VDKLKPMNIE RKQTRKEYNT SILEFRWLTS SKSVIVSQFC AFDSSSNTYR SRAQQVPPYG VYHPPFIKYA CL AIRKNGQ IDFWYQFSNS KDHKKITLQL LDTSNQRFKD LQWLEFARIT PMNDDQCMLI TTYSKLSKNI SFYKLHVNWN LNA TKPNVL NDPSLKIQFI LSTTLDPTDD EGHVLKLENL HVVSKSSIEK DPSPEILVLY NVCDTSKSLV KRYRLAPTQL SAEY LVILK PDLNIDRNNS TNQIFQSRRY NLRRHSDIVL DKKVTLITSE MFDAFVSFYF EDGTIESYNQ NDWKLETERL ISQSQ LGKF KNIIASPLSA GFNYGKLPLP PSVEWMKVSP SMCGVIVKQY NKKWPQFYAA VQKNYADPEK DSINATALAF GYVKSL HKQ ISAEDLTIAA KTHILRISFL DRKRAKEFIT TLLKSLYSFF NISPDAPKEI MDKIITSRPL QKIMLLQLEL GSCFSQE NI EEMARVILYL KNVLFAFNGV ARNFHFAIEQ ISNNSNQQQN PKLFQTIFSK QDLIHSLIPV AKWFVKFITY LTQEILIL I NDPTNKEYTL VHGIFGAKMS RTLILSILNE IKKVTQIVAK FPETSYPILN ESSTFLKLVL SESPVDFEKF ETFLVDVNN KFIALCEQQP SQEREFSLLV KAEIPPEYAK VGDFLLQYAN NAVISHANAA AVYFADTSGL KISNSEFFNP EIFHLLQPLE EGLIIDTDK LPIKNRTSKS FSKLLYDDVT CDKLSVSEIS DGKLKRCSRC GSVTRAGNII SSDKTIVPTS IQTKRWPTMY T RLCICSGM LFEMDG |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | .1 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
| |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM CPC |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.0 µm / Calibrated defocus min: 0.5 µm / Calibrated magnification: 64000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 64000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 64000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 40900 / Average exposure time: 3.4 sec. / Average electron dose: 42.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z
Z Y
Y X
X










































