+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7uik | ||||||
|---|---|---|---|---|---|---|---|
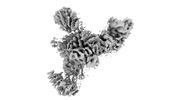
| Title | Mediator-PIC Early (Tail A + Upstream DNA & Activator) | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  TRANSCRIPTION / Divergent transcription / Mediator / TRANSCRIPTION / Divergent transcription / Mediator /  RNA Polymerase II / PIC / Pre-Initiation / RNA Polymerase II / PIC / Pre-Initiation /  Activation / Activation /  Transcription Factor / Gal4 / Gal4VP16 Transcription Factor / Gal4 / Gal4VP16 | ||||||
| Function / homology |  Function and homology information Function and homology information regulation of transcription from RNA polymerase II promoter by galactose / TFIIE-class transcription factor complex binding / regulation of establishment of protein localization to chromosome / positive regulation of invasive growth in response to glucose limitation / TFIIH-class transcription factor complex binding / core mediator complex / galactose metabolic process / regulation of transcription from RNA polymerase II promoter by galactose / TFIIE-class transcription factor complex binding / regulation of establishment of protein localization to chromosome / positive regulation of invasive growth in response to glucose limitation / TFIIH-class transcription factor complex binding / core mediator complex / galactose metabolic process /  mediator complex / positive regulation of transcription from RNA polymerase II promoter by galactose / TFIIB-class transcription factor binding ... mediator complex / positive regulation of transcription from RNA polymerase II promoter by galactose / TFIIB-class transcription factor binding ... regulation of transcription from RNA polymerase II promoter by galactose / TFIIE-class transcription factor complex binding / regulation of establishment of protein localization to chromosome / positive regulation of invasive growth in response to glucose limitation / TFIIH-class transcription factor complex binding / core mediator complex / galactose metabolic process / regulation of transcription from RNA polymerase II promoter by galactose / TFIIE-class transcription factor complex binding / regulation of establishment of protein localization to chromosome / positive regulation of invasive growth in response to glucose limitation / TFIIH-class transcription factor complex binding / core mediator complex / galactose metabolic process /  mediator complex / positive regulation of transcription from RNA polymerase II promoter by galactose / TFIIB-class transcription factor binding / positive regulation of transcription initiation by RNA polymerase II / RNA polymerase II core promoter sequence-specific DNA binding / RNA polymerase II preinitiation complex assembly / transcription repressor complex / positive regulation of transcription elongation by RNA polymerase II / mediator complex / positive regulation of transcription from RNA polymerase II promoter by galactose / TFIIB-class transcription factor binding / positive regulation of transcription initiation by RNA polymerase II / RNA polymerase II core promoter sequence-specific DNA binding / RNA polymerase II preinitiation complex assembly / transcription repressor complex / positive regulation of transcription elongation by RNA polymerase II /  transcription coregulator activity / DNA-binding transcription activator activity, RNA polymerase II-specific / RNA polymerase II-specific DNA-binding transcription factor binding / transcription by RNA polymerase II / transcription coregulator activity / DNA-binding transcription activator activity, RNA polymerase II-specific / RNA polymerase II-specific DNA-binding transcription factor binding / transcription by RNA polymerase II /  transcription coactivator activity / DNA-binding transcription factor activity, RNA polymerase II-specific / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-templated transcription / regulation of transcription by RNA polymerase II / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / transcription coactivator activity / DNA-binding transcription factor activity, RNA polymerase II-specific / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-templated transcription / regulation of transcription by RNA polymerase II / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II /  mitochondrion / zinc ion binding / identical protein binding / mitochondrion / zinc ion binding / identical protein binding /  nucleus / nucleus /  cytosol cytosolSimilarity search - Function | ||||||
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast)  Saccharomyces cerevisiae S288C (yeast) Saccharomyces cerevisiae S288C (yeast) | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 7.7 Å cryo EM / Resolution: 7.7 Å | ||||||
 Authors Authors | Gorbea Colon, J.J. / Chen, S.-F. / Tsai, K.L. / Murakami, K. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: Mol Cell / Year: 2023 Journal: Mol Cell / Year: 2023Title: Structural basis of a transcription pre-initiation complex on a divergent promoter. Authors: Jose J Gorbea Colón / Leon Palao / Shin-Fu Chen / Hee Jong Kim / Laura Snyder / Yi-Wei Chang / Kuang-Lei Tsai / Kenji Murakami /  Abstract: Most eukaryotic promoter regions are divergently transcribed. As the RNA polymerase II pre-initiation complex (PIC) is intrinsically asymmetric and responsible for transcription in a single ...Most eukaryotic promoter regions are divergently transcribed. As the RNA polymerase II pre-initiation complex (PIC) is intrinsically asymmetric and responsible for transcription in a single direction, it is unknown how divergent transcription arises. Here, the Saccharomyces cerevisiae Mediator complexed with a PIC (Med-PIC) was assembled on a divergent promoter and analyzed by cryoelectron microscopy. The structure reveals two distinct Med-PICs forming a dimer through the Mediator tail module, induced by a homodimeric activator protein localized near the dimerization interface. The tail dimer is associated with ∼80-bp upstream DNA, such that two flanking core promoter regions are positioned and oriented in a suitable form for PIC assembly in opposite directions. Also, cryoelectron tomography visualized the progress of the PIC assembly on the two core promoter regions, providing direct evidence for the role of the Med-PIC dimer in divergent transcription. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7uik.cif.gz 7uik.cif.gz | 507.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7uik.ent.gz pdb7uik.ent.gz | 380.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7uik.json.gz 7uik.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ui/7uik https://data.pdbj.org/pub/pdb/validation_reports/ui/7uik ftp://data.pdbj.org/pub/pdb/validation_reports/ui/7uik ftp://data.pdbj.org/pub/pdb/validation_reports/ui/7uik | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  26547MC  7ui9C  7uicC  7uifC  7uigC  7uilC  7uioC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-DNA chain , 2 types, 2 molecules XY
| #1: DNA chain | Mass: 11666.479 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast)Production host:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
|---|---|
| #2: DNA chain | Mass: 11424.310 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast)Production host:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Mediator of RNA polymerase II transcription subunit ... , 6 types, 6 molecules obcnep
| #4: Protein | Mass: 120432.242 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Saccharomyces cerevisiae S288C (yeast) / Strain: ATCC 204508 / S288c / References: UniProt: P19659 Saccharomyces cerevisiae S288C (yeast) / Strain: ATCC 204508 / S288c / References: UniProt: P19659 |
|---|---|
| #5: Protein | Mass: 47754.270 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Saccharomyces cerevisiae S288C (yeast) / Strain: ATCC 204508 / S288c / References: UniProt: Q12124 Saccharomyces cerevisiae S288C (yeast) / Strain: ATCC 204508 / S288c / References: UniProt: Q12124 |
| #6: Protein | Mass: 43116.039 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Saccharomyces cerevisiae S288C (yeast) / References: UniProt: P40356 Saccharomyces cerevisiae S288C (yeast) / References: UniProt: P40356 |
| #7: Protein | Mass: 27535.459 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Saccharomyces cerevisiae S288C (yeast) / References: UniProt: P19263 Saccharomyces cerevisiae S288C (yeast) / References: UniProt: P19263 |
| #8: Protein | Mass: 128918.750 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Saccharomyces cerevisiae S288C (yeast) / Strain: ATCC 204508 / S288c / References: UniProt: P53114 Saccharomyces cerevisiae S288C (yeast) / Strain: ATCC 204508 / S288c / References: UniProt: P53114 |
| #9: Protein | Mass: 111423.422 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Saccharomyces cerevisiae S288C (yeast) / Strain: ATCC 204508 / S288c / References: UniProt: P32259 Saccharomyces cerevisiae S288C (yeast) / Strain: ATCC 204508 / S288c / References: UniProt: P32259 |
-Protein / Non-polymers , 2 types, 6 molecules TU

| #10: Chemical | ChemComp-ZN / #3: Protein | Mass: 16895.672 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Saccharomyces cerevisiae S288C (yeast) / Gene: GAL4, YPL248C / Production host: Saccharomyces cerevisiae S288C (yeast) / Gene: GAL4, YPL248C / Production host:   Escherichia coli (E. coli) / References: UniProt: P04386 Escherichia coli (E. coli) / References: UniProt: P04386 |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Med-PICEarly (TailA+Activator & DNA Class 1) / Type: COMPLEX / Entity ID: #1-#2, #4-#6, #3, #8-#9 / Source: MULTIPLE SOURCES | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Experimental value: NO | |||||||||||||||||||||||||
| Source (natural) | Organism:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) | |||||||||||||||||||||||||
| Source (recombinant) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) | |||||||||||||||||||||||||
| Buffer solution | pH: 7.6 | |||||||||||||||||||||||||
| Buffer component |
| |||||||||||||||||||||||||
| Specimen | Conc.: 0.1 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES | |||||||||||||||||||||||||
Vitrification | Instrument: LEICA EM CPC / Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal magnification: 64000 X / Calibrated magnification: 64000 X / Nominal defocus max: 3000 nm / Nominal defocus min: 500 nm / Calibrated defocus min: 500 nm / Calibrated defocus max: 3000 nm / Cs Bright-field microscopy / Nominal magnification: 64000 X / Calibrated magnification: 64000 X / Nominal defocus max: 3000 nm / Nominal defocus min: 500 nm / Calibrated defocus min: 500 nm / Calibrated defocus max: 3000 nm / Cs : 2.7 mm : 2.7 mm |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 3.4 sec. / Electron dose: 42 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) / Num. of real images: 40900 |
- Processing
Processing
CTF correction | Type: NONE |
|---|---|
3D reconstruction | Resolution: 7.7 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 199834 / Symmetry type: POINT |
 Movie
Movie Controller
Controller









 PDBj
PDBj







































