+ Open data
Open data
- Basic information
Basic information
| Entry | 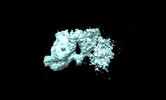 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Dimerized Mediator PIC with early GTFs on a divergent promoter | |||||||||
 Map data Map data | Dimerized Mediator PIC with early GTFs on a divergent promoter | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) | |||||||||
| Method | subtomogram averaging /  cryo EM / Resolution: 36.0 Å cryo EM / Resolution: 36.0 Å | |||||||||
 Authors Authors | Palao III L | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2023 Journal: Mol Cell / Year: 2023Title: Structural basis of a transcription pre-initiation complex on a divergent promoter. Authors: Jose J Gorbea Colón / Leon Palao / Shin-Fu Chen / Hee Jong Kim / Laura Snyder / Yi-Wei Chang / Kuang-Lei Tsai / Kenji Murakami /  Abstract: Most eukaryotic promoter regions are divergently transcribed. As the RNA polymerase II pre-initiation complex (PIC) is intrinsically asymmetric and responsible for transcription in a single ...Most eukaryotic promoter regions are divergently transcribed. As the RNA polymerase II pre-initiation complex (PIC) is intrinsically asymmetric and responsible for transcription in a single direction, it is unknown how divergent transcription arises. Here, the Saccharomyces cerevisiae Mediator complexed with a PIC (Med-PIC) was assembled on a divergent promoter and analyzed by cryoelectron microscopy. The structure reveals two distinct Med-PICs forming a dimer through the Mediator tail module, induced by a homodimeric activator protein localized near the dimerization interface. The tail dimer is associated with ∼80-bp upstream DNA, such that two flanking core promoter regions are positioned and oriented in a suitable form for PIC assembly in opposite directions. Also, cryoelectron tomography visualized the progress of the PIC assembly on the two core promoter regions, providing direct evidence for the role of the Med-PIC dimer in divergent transcription. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28024.map.gz emd_28024.map.gz | 15.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28024-v30.xml emd-28024-v30.xml emd-28024.xml emd-28024.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28024_fsc.xml emd_28024_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_28024.png emd_28024.png | 36.4 KB | ||
| Others |  emd_28024_half_map_1.map.gz emd_28024_half_map_1.map.gz emd_28024_half_map_2.map.gz emd_28024_half_map_2.map.gz | 15.8 MB 15.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28024 http://ftp.pdbj.org/pub/emdb/structures/EMD-28024 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28024 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28024 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28024.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28024.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Dimerized Mediator PIC with early GTFs on a divergent promoter | ||||||||||||||||||||
| Voxel size | X=Y=Z: 3.352 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 1
| File | emd_28024_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 2
| File | emd_28024_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of Mediator, RNA Polymerase II, and general transcription...
| Entire | Name: Complex of Mediator, RNA Polymerase II, and general transcription factors (TFIIA, TFIIB, TBP, TFIIF, TFIIS) on a divergent promoter. |
|---|---|
| Components |
|
-Supramolecule #1: Complex of Mediator, RNA Polymerase II, and general transcription...
| Supramolecule | Name: Complex of Mediator, RNA Polymerase II, and general transcription factors (TFIIA, TFIIB, TBP, TFIIF, TFIIS) on a divergent promoter. type: complex / ID: 1 / Chimera: Yes / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
Details: 20mM HEPES pH7.6, 2mM Magnesium Acetate, 80mM Potassium Acetate, 10nm gold beads | ||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 65 % / Chamber temperature: 298 K / Instrument: LEICA EM CPC | ||||||||||||
| Details | Samples were chemically stabilized by fixation in glycerol gradients with increasing concentrations of glutaraldehyde (0-0.125%). Peak gradient fractions containing glutaraldhyde were quenched upon the addition of 50 mM glycine (pH 7.6) and 10nm gold beads were added before flash-freezing and storage under liquid nitrogen. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 0.3 µm / Nominal magnification: 53000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 0.3 µm / Nominal magnification: 53000 |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 1.95 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z
Z Y
Y X
X


















