+Search query
-Structure paper
| Title | High-Resolution Structural Proteomics of Mitochondria Using the 'Build and Retrieve' Methodology. |
|---|---|
| Journal, issue, pages | Mol Cell Proteomics, Vol. 22, Issue 12, Page 100666, Year 2023 |
| Publish date | Oct 14, 2023 |
 Authors Authors | Zhemin Zhang / Marios L Tringides / Christopher E Morgan / Masaru Miyagi / Jason A Mears / Charles L Hoppel / Edward W Yu /  |
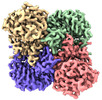
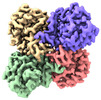
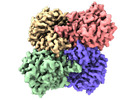
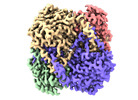
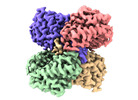
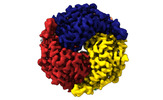
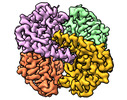
| PubMed Abstract | The application of integrated systems biology to the field of structural biology is a promising new direction, although it is still in the infant stages of development. Here we report the use of ...The application of integrated systems biology to the field of structural biology is a promising new direction, although it is still in the infant stages of development. Here we report the use of single particle cryo-EM to identify multiple proteins from three enriched heterogeneous fractions prepared from human liver mitochondrial lysate. We simultaneously identify and solve high-resolution structures of nine essential mitochondrial enzymes with key metabolic functions, including fatty acid catabolism, reactive oxidative species clearance, and amino acid metabolism. Our methodology also identified multiple distinct members of the acyl-CoA dehydrogenase family. This work highlights the potential of cryo-EM to explore tissue proteomics at the atomic level. |
 External links External links |  Mol Cell Proteomics / Mol Cell Proteomics /  PubMed:37839702 / PubMed:37839702 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.31 - 3.15 Å |
| Structure data | EMDB-40463, PDB-8sgp: EMDB-40465, PDB-8sgr: EMDB-40466, PDB-8sgs: EMDB-40469, PDB-8sgv: EMDB-40493, PDB-8shs: EMDB-40556, PDB-8sk6: EMDB-40558, PDB-8sk8: EMDB-40565, PDB-8skr: EMDB-40566, PDB-8sks: |
| Chemicals |  ChemComp-FAD:  ChemComp-COA:  ChemComp-HEM:  ChemComp-NDP:  ChemComp-PLP:  ChemComp-MN: |
| Source |
|
 Keywords Keywords |  OXIDOREDUCTASE / OXIDOREDUCTASE /  human / human /  liver / liver /  mitochondrial / Medium-chain specific acyl-CoA dehydrogenase / mitochondrial / Medium-chain specific acyl-CoA dehydrogenase /  Isovaleryl-CoA dehydrogenase / Short-chain specific acyl-CoA dehydrogenase / Isovaleryl-CoA dehydrogenase / Short-chain specific acyl-CoA dehydrogenase /  Catalase / Catalase /  Aldehyde dehydrogenase / ALDH2 / Aldehyde dehydrogenase / ALDH2 /  ISOMERASE / Delta(3 / 5)-Delta(2 / 4)-dienoyl-CoA isomerase / ISOMERASE / Delta(3 / 5)-Delta(2 / 4)-dienoyl-CoA isomerase /  Glutamate dehydrogenase 1 / Glutamate dehydrogenase 1 /  TRANSFERASE / TRANSFERASE /  Aspartate aminotransferase / Superoxide dismutase [Mn] Aspartate aminotransferase / Superoxide dismutase [Mn] |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers