[English] 日本語
 Yorodumi
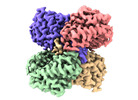
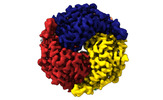
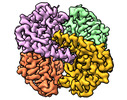
Yorodumi- PDB-8sgp: human liver mitochondrial Medium-chain specific acyl-CoA dehydrogenase -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8sgp | ||||||
|---|---|---|---|---|---|---|---|
| Title | human liver mitochondrial Medium-chain specific acyl-CoA dehydrogenase | ||||||
 Components Components | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial | ||||||
 Keywords Keywords |  OXIDOREDUCTASE / OXIDOREDUCTASE /  human / human /  liver / liver /  mitochondrial / Medium-chain specific acyl-CoA dehydrogenase mitochondrial / Medium-chain specific acyl-CoA dehydrogenase | ||||||
| Function / homology |  Function and homology information Function and homology informationmedium-chain fatty acid catabolic process / mitochondrial fatty acid beta-oxidation of unsaturated fatty acids / carnitine metabolic process, CoA-linked /  medium-chain acyl-CoA dehydrogenase / Beta oxidation of octanoyl-CoA to hexanoyl-CoA / Beta oxidation of decanoyl-CoA to octanoyl-CoA-CoA / medium-chain fatty acyl-CoA dehydrogenase activity / carnitine biosynthetic process / fatty acid beta-oxidation using acyl-CoA dehydrogenase / medium-chain fatty acid metabolic process ...medium-chain fatty acid catabolic process / mitochondrial fatty acid beta-oxidation of unsaturated fatty acids / carnitine metabolic process, CoA-linked / medium-chain acyl-CoA dehydrogenase / Beta oxidation of octanoyl-CoA to hexanoyl-CoA / Beta oxidation of decanoyl-CoA to octanoyl-CoA-CoA / medium-chain fatty acyl-CoA dehydrogenase activity / carnitine biosynthetic process / fatty acid beta-oxidation using acyl-CoA dehydrogenase / medium-chain fatty acid metabolic process ...medium-chain fatty acid catabolic process / mitochondrial fatty acid beta-oxidation of unsaturated fatty acids / carnitine metabolic process, CoA-linked /  medium-chain acyl-CoA dehydrogenase / Beta oxidation of octanoyl-CoA to hexanoyl-CoA / Beta oxidation of decanoyl-CoA to octanoyl-CoA-CoA / medium-chain fatty acyl-CoA dehydrogenase activity / carnitine biosynthetic process / fatty acid beta-oxidation using acyl-CoA dehydrogenase / medium-chain fatty acid metabolic process / medium-chain acyl-CoA dehydrogenase / Beta oxidation of octanoyl-CoA to hexanoyl-CoA / Beta oxidation of decanoyl-CoA to octanoyl-CoA-CoA / medium-chain fatty acyl-CoA dehydrogenase activity / carnitine biosynthetic process / fatty acid beta-oxidation using acyl-CoA dehydrogenase / medium-chain fatty acid metabolic process /  acyl-CoA dehydrogenase activity / cardiac muscle cell differentiation / glycogen biosynthetic process / acyl-CoA dehydrogenase activity / cardiac muscle cell differentiation / glycogen biosynthetic process /  regulation of gluconeogenesis / fatty acid beta-oxidation / response to starvation / response to cold / post-embryonic development / liver development / regulation of gluconeogenesis / fatty acid beta-oxidation / response to starvation / response to cold / post-embryonic development / liver development /  mitochondrial membrane / PPARA activates gene expression / mitochondrial membrane / PPARA activates gene expression /  flavin adenine dinucleotide binding / flavin adenine dinucleotide binding /  mitochondrial matrix / mitochondrial matrix /  axon / axon /  mitochondrion / identical protein binding / mitochondrion / identical protein binding /  nucleus / nucleus /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.69 Å cryo EM / Resolution: 2.69 Å | ||||||
 Authors Authors | Zhang, Z. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: Mol Cell Proteomics / Year: 2023 Journal: Mol Cell Proteomics / Year: 2023Title: High-Resolution Structural Proteomics of Mitochondria Using the 'Build and Retrieve' Methodology. Authors: Zhemin Zhang / Marios L Tringides / Christopher E Morgan / Masaru Miyagi / Jason A Mears / Charles L Hoppel / Edward W Yu /  Abstract: The application of integrated systems biology to the field of structural biology is a promising new direction, although it is still in the infant stages of development. Here we report the use of ...The application of integrated systems biology to the field of structural biology is a promising new direction, although it is still in the infant stages of development. Here we report the use of single particle cryo-EM to identify multiple proteins from three enriched heterogeneous fractions prepared from human liver mitochondrial lysate. We simultaneously identify and solve high-resolution structures of nine essential mitochondrial enzymes with key metabolic functions, including fatty acid catabolism, reactive oxidative species clearance, and amino acid metabolism. Our methodology also identified multiple distinct members of the acyl-CoA dehydrogenase family. This work highlights the potential of cryo-EM to explore tissue proteomics at the atomic level. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8sgp.cif.gz 8sgp.cif.gz | 261.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8sgp.ent.gz pdb8sgp.ent.gz | 211.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8sgp.json.gz 8sgp.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/sg/8sgp https://data.pdbj.org/pub/pdb/validation_reports/sg/8sgp ftp://data.pdbj.org/pub/pdb/validation_reports/sg/8sgp ftp://data.pdbj.org/pub/pdb/validation_reports/sg/8sgp | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  40463MC  8sgrC  8sgsC  8sgvC  8shsC  8sk6C  8sk8C  8skrC  8sksC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 46648.273 Da / Num. of mol.: 4 / Source method: isolated from a natural source / Source: (natural)   Homo sapiens (human) Homo sapiens (human)References: UniProt: P11310,  medium-chain acyl-CoA dehydrogenase medium-chain acyl-CoA dehydrogenase#2: Chemical | ChemComp-FAD /  Flavin adenine dinucleotide Flavin adenine dinucleotideHas ligand of interest | N | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Medium-chain specific acyl-CoA dehydrogenase / Type: COMPLEX / Entity ID: #1 / Source: NATURAL |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Source (recombinant) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Buffer solution | pH: 7.5 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES |
Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal magnification: 105000 X / Nominal defocus max: 1500 nm / Nominal defocus min: 800 nm Bright-field microscopy / Nominal magnification: 105000 X / Nominal defocus max: 1500 nm / Nominal defocus min: 800 nm |
| Image recording | Electron dose: 35 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.20.1_4487: / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
3D reconstruction | Resolution: 2.69 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 18616 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller




 PDBj
PDBj

