+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
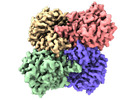
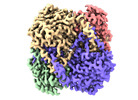
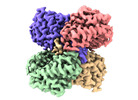
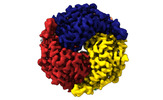
| Title | human liver mitochondrial Aspartate aminotransferase | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  human / human /  liver / liver /  mitochondrial / mitochondrial /  Aspartate aminotransferase / Aspartate aminotransferase /  TRANSFERASE TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology information4-hydroxyproline catabolic process / glutamate catabolic process to aspartate / glutamate catabolic process to 2-oxoglutarate / aspartate biosynthetic process / Degradation of cysteine and homocysteine / aspartate catabolic process /  kynurenine-oxoglutarate transaminase / kynurenine-oxoglutarate transaminase /  kynurenine-oxoglutarate transaminase activity / aspartate metabolic process / glutamate metabolic process ...4-hydroxyproline catabolic process / glutamate catabolic process to aspartate / glutamate catabolic process to 2-oxoglutarate / aspartate biosynthetic process / Degradation of cysteine and homocysteine / aspartate catabolic process / kynurenine-oxoglutarate transaminase activity / aspartate metabolic process / glutamate metabolic process ...4-hydroxyproline catabolic process / glutamate catabolic process to aspartate / glutamate catabolic process to 2-oxoglutarate / aspartate biosynthetic process / Degradation of cysteine and homocysteine / aspartate catabolic process /  kynurenine-oxoglutarate transaminase / kynurenine-oxoglutarate transaminase /  kynurenine-oxoglutarate transaminase activity / aspartate metabolic process / glutamate metabolic process / Aspartate and asparagine metabolism / Glutamate and glutamine metabolism / 2-oxoglutarate metabolic process / oxaloacetate metabolic process / kynurenine-oxoglutarate transaminase activity / aspartate metabolic process / glutamate metabolic process / Aspartate and asparagine metabolism / Glutamate and glutamine metabolism / 2-oxoglutarate metabolic process / oxaloacetate metabolic process /  aspartate transaminase / aspartate transaminase /  L-aspartate:2-oxoglutarate aminotransferase activity / Glyoxylate metabolism and glycine degradation / L-aspartate:2-oxoglutarate aminotransferase activity / Glyoxylate metabolism and glycine degradation /  Gluconeogenesis / fatty acid transport / Gluconeogenesis / fatty acid transport /  pyridoxal phosphate binding / response to ethanol / pyridoxal phosphate binding / response to ethanol /  mitochondrial matrix / mitochondrial matrix /  mitochondrion / mitochondrion /  RNA binding / extracellular exosome / RNA binding / extracellular exosome /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.99 Å cryo EM / Resolution: 2.99 Å | |||||||||
 Authors Authors | Zhang Z / Tringides M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell Proteomics / Year: 2023 Journal: Mol Cell Proteomics / Year: 2023Title: High-Resolution Structural Proteomics of Mitochondria Using the 'Build and Retrieve' Methodology. Authors: Zhemin Zhang / Marios L Tringides / Christopher E Morgan / Masaru Miyagi / Jason A Mears / Charles L Hoppel / Edward W Yu /  Abstract: The application of integrated systems biology to the field of structural biology is a promising new direction, although it is still in the infant stages of development. Here we report the use of ...The application of integrated systems biology to the field of structural biology is a promising new direction, although it is still in the infant stages of development. Here we report the use of single particle cryo-EM to identify multiple proteins from three enriched heterogeneous fractions prepared from human liver mitochondrial lysate. We simultaneously identify and solve high-resolution structures of nine essential mitochondrial enzymes with key metabolic functions, including fatty acid catabolism, reactive oxidative species clearance, and amino acid metabolism. Our methodology also identified multiple distinct members of the acyl-CoA dehydrogenase family. This work highlights the potential of cryo-EM to explore tissue proteomics at the atomic level. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40565.map.gz emd_40565.map.gz | 51.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40565-v30.xml emd-40565-v30.xml emd-40565.xml emd-40565.xml | 13.4 KB 13.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40565_fsc.xml emd_40565_fsc.xml | 11.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_40565.png emd_40565.png | 97.8 KB | ||
| Filedesc metadata |  emd-40565.cif.gz emd-40565.cif.gz | 5.1 KB | ||
| Others |  emd_40565_half_map_1.map.gz emd_40565_half_map_1.map.gz emd_40565_half_map_2.map.gz emd_40565_half_map_2.map.gz | 95.3 MB 95.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40565 http://ftp.pdbj.org/pub/emdb/structures/EMD-40565 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40565 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40565 | HTTPS FTP |
-Related structure data
| Related structure data |  8skrMC  8sgpC  8sgrC  8sgsC  8sgvC  8shsC  8sk6C  8sk8C  8sksC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40565.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40565.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_40565_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_40565_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Aspartate aminotransferase
| Entire | Name: Aspartate aminotransferase Aspartate transaminase Aspartate transaminase |
|---|---|
| Components |
|
-Supramolecule #1: Aspartate aminotransferase
| Supramolecule | Name: Aspartate aminotransferase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Aspartate aminotransferase, mitochondrial
| Macromolecule | Name: Aspartate aminotransferase, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number:  aspartate transaminase aspartate transaminase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 47.580578 KDa |
| Sequence | String: MALLHSGRVL PGIAAAFHPG LAAAASARAS SWWTHVEMGP PDPILGVTEA FKRDTNSKKM NLGVGAYRDD NGKPYVLPSV RKAEAQIAA KNLDKEYLPI GGLAEFCKAS AELALGENSE VLKSGRFVTV QTISGTGALR IGASFLQRFF KFSRDVFLPK P TWGNHTPI ...String: MALLHSGRVL PGIAAAFHPG LAAAASARAS SWWTHVEMGP PDPILGVTEA FKRDTNSKKM NLGVGAYRDD NGKPYVLPSV RKAEAQIAA KNLDKEYLPI GGLAEFCKAS AELALGENSE VLKSGRFVTV QTISGTGALR IGASFLQRFF KFSRDVFLPK P TWGNHTPI FRDAGMQLQG YRYYDPKTCG FDFTGAVEDI SKIPEQSVLL LHACAHNPTG VDPRPEQWKE IATVVKKRNL FA FFDMAYQ GFASGDGDKD AWAVRHFIEQ GINVCLCQSY AKNMGLYGER VGAFTMVCKD ADEAKRVESQ LKILIRPMYS NPP LNGARI AAAILNTPDL RKQWLQEVKV MADRIIGMRT QLVSNLKKEG STHNWQHITD QIGMFCFTGL KPEQVERLIK EFSI YMTKD GRISVAGVTS SNVGYLAHAI HQVTK UniProtKB:  Aspartate aminotransferase, mitochondrial Aspartate aminotransferase, mitochondrial |
-Macromolecule #2: PYRIDOXAL-5'-PHOSPHATE
| Macromolecule | Name: PYRIDOXAL-5'-PHOSPHATE / type: ligand / ID: 2 / Number of copies: 2 / Formula: PLP |
|---|---|
| Molecular weight | Theoretical: 247.142 Da |
| Chemical component information |  ChemComp-PLP: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 Bright-field microscopy / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 35.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





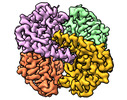

 Z
Z Y
Y X
X