+Search query
-Structure paper
| Title | Structure, folding and flexibility of co-transcriptional RNA origami. |
|---|---|
| Journal, issue, pages | Nat Nanotechnol, Vol. 18, Issue 7, Page 808-817, Year 2023 |
| Publish date | Feb 27, 2023 |
 Authors Authors | Ewan K S McRae / Helena Østergaard Rasmussen / Jianfang Liu / Andreas Bøggild / Michael T A Nguyen / Nestor Sampedro Vallina / Thomas Boesen / Jan Skov Pedersen / Gang Ren / Cody Geary / Ebbe Sloth Andersen /   |
| PubMed Abstract | RNA origami is a method for designing RNA nanostructures that can self-assemble through co-transcriptional folding with applications in nanomedicine and synthetic biology. However, to advance the ...RNA origami is a method for designing RNA nanostructures that can self-assemble through co-transcriptional folding with applications in nanomedicine and synthetic biology. However, to advance the method further, an improved understanding of RNA structural properties and folding principles is required. Here we use cryogenic electron microscopy to study RNA origami sheets and bundles at sub-nanometre resolution revealing structural parameters of kissing-loop and crossover motifs, which are used to improve designs. In RNA bundle designs, we discover a kinetic folding trap that forms during folding and is only released after 10 h. Exploration of the conformational landscape of several RNA designs reveal the flexibility of helices and structural motifs. Finally, sheets and bundles are combined to construct a multidomain satellite shape, which is characterized by individual-particle cryo-electron tomography to reveal the domain flexibility. Together, the study provides a structural basis for future improvements to the design cycle of genetically encoded RNA nanodevices. |
 External links External links |  Nat Nanotechnol / Nat Nanotechnol /  PubMed:36849548 / PubMed:36849548 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / EM (tomography) |
| Resolution | 4.08 - 27.0 Å |
| Structure data |  EMDB-13592: Cryo-EM SPA reconstruction of an extended RNA origami 5 helix tile (5HT-B-3X)  EMDB-13625: Mature conformer 2 of a 6-helix bundle of RNA with a clasp  EMDB-13626: Young conformer 2 of a 6-helix bundle of RNA with a clasp  EMDB-13627: 6-Helix bundle of RNA EMDB-13628, PDB-7ptk: EMDB-13630, PDB-7ptl: EMDB-13633, PDB-7ptq: EMDB-13636, PDB-7pts: EMDB-13926, PDB-7qdu:  EMDB-28669: Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 01) 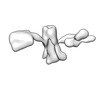 EMDB-28670: Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 02) 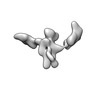 EMDB-28671: Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 03) 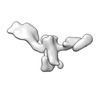 EMDB-28672: Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 04)  EMDB-28673: Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 05) 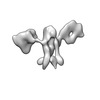 EMDB-28674: Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 06) 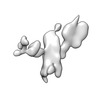 EMDB-28675: Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 07) 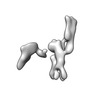 EMDB-28676: Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 08)  EMDB-28677: Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 09)  EMDB-28678: Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 10)  EMDB-28679: Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 11)  EMDB-28680: Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 12)  EMDB-28681: Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 13)  EMDB-28682: Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 14) 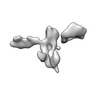 EMDB-28683: Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 15) 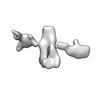 EMDB-28684: Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 16) |
| Source |
|
 Keywords Keywords |  RNA / RNA /  origami / origami /  nanostructure nanostructure |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers













