+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7qdu | ||||||
|---|---|---|---|---|---|---|---|
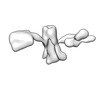
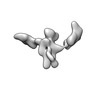
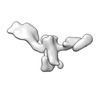
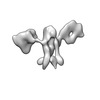
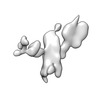
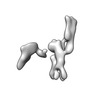
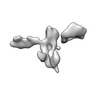
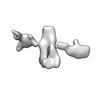
| Title | Twist-corrected RNA origami 5-helix Tile A | ||||||
 Components Components | Chains: Q | ||||||
 Keywords Keywords |  RNA / RNA /  Origami / Origami /  nanostructure nanostructure | ||||||
| Function / homology |  RNA / RNA (> 10) / RNA (> 100) RNA / RNA (> 10) / RNA (> 100) Function and homology information Function and homology information | ||||||
| Biological species | synthetic construct (others) | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 5.14 Å cryo EM / Resolution: 5.14 Å | ||||||
 Authors Authors | McRae, E.K.S. / Andersen, E.S. | ||||||
| Funding support |  Denmark, 1items Denmark, 1items
| ||||||
 Citation Citation |  Journal: Nat Nanotechnol / Year: 2023 Journal: Nat Nanotechnol / Year: 2023Title: Structure, folding and flexibility of co-transcriptional RNA origami. Authors: Ewan K S McRae / Helena Østergaard Rasmussen / Jianfang Liu / Andreas Bøggild / Michael T A Nguyen / Nestor Sampedro Vallina / Thomas Boesen / Jan Skov Pedersen / Gang Ren / Cody Geary / ...Authors: Ewan K S McRae / Helena Østergaard Rasmussen / Jianfang Liu / Andreas Bøggild / Michael T A Nguyen / Nestor Sampedro Vallina / Thomas Boesen / Jan Skov Pedersen / Gang Ren / Cody Geary / Ebbe Sloth Andersen /   Abstract: RNA origami is a method for designing RNA nanostructures that can self-assemble through co-transcriptional folding with applications in nanomedicine and synthetic biology. However, to advance the ...RNA origami is a method for designing RNA nanostructures that can self-assemble through co-transcriptional folding with applications in nanomedicine and synthetic biology. However, to advance the method further, an improved understanding of RNA structural properties and folding principles is required. Here we use cryogenic electron microscopy to study RNA origami sheets and bundles at sub-nanometre resolution revealing structural parameters of kissing-loop and crossover motifs, which are used to improve designs. In RNA bundle designs, we discover a kinetic folding trap that forms during folding and is only released after 10 h. Exploration of the conformational landscape of several RNA designs reveal the flexibility of helices and structural motifs. Finally, sheets and bundles are combined to construct a multidomain satellite shape, which is characterized by individual-particle cryo-electron tomography to reveal the domain flexibility. Together, the study provides a structural basis for future improvements to the design cycle of genetically encoded RNA nanodevices. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7qdu.cif.gz 7qdu.cif.gz | 402.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7qdu.ent.gz pdb7qdu.ent.gz | 298.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7qdu.json.gz 7qdu.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qd/7qdu https://data.pdbj.org/pub/pdb/validation_reports/qd/7qdu ftp://data.pdbj.org/pub/pdb/validation_reports/qd/7qdu ftp://data.pdbj.org/pub/pdb/validation_reports/qd/7qdu | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  13926MC  7ptkC  7ptlC  7ptqC  7ptsC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: RNA chain | Mass: 177720.031 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: In vitro transcribed RNA / Source: (synth.) synthetic construct (others) |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Twist-corrected 5-helix tile A. / Type: COMPLEX / Details: In vitro transcribed RNA purified by SEC. / Entity ID: all / Source: RECOMBINANT | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Value: 0.175 MDa / Experimental value: NO | ||||||||||||||||||||
| Source (natural) | Organism: synthetic construct (others) | ||||||||||||||||||||
| Source (recombinant) | Organism: synthetic construct (others) | ||||||||||||||||||||
| Buffer solution | pH: 8 Details: Freshly prepared and filtered through 0.22 micron filter prior to use. | ||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||
| Specimen | Conc.: 3 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YESDetails: Sample was purified by size exclusion chromatography and concentrated in an Amicon spin concentrator. | ||||||||||||||||||||
| Specimen support | Details: 15mA current on a glocube. / Grid material: GOLD / Grid mesh size: 300 divisions/in. / Grid type: UltrAuFoil R1.2/1.3 | ||||||||||||||||||||
Vitrification | Instrument: LEICA EM GP / Cryogen name: ETHANE / Humidity: 99 % / Chamber temperature: 294 K Details: 3 microlitre droplet, 4 second delay before blotting, 6 second blot, 0 second delay before plunging. |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal magnification: 130000 X / Cs Bright-field microscopy / Nominal magnification: 130000 X / Cs : 2.7 mm / C2 aperture diameter: 50 µm / Alignment procedure: COMA FREE : 2.7 mm / C2 aperture diameter: 50 µm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 1.5 sec. / Electron dose: 60 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Num. of grids imaged: 1 / Num. of real images: 6122 / Details: Images were collected as 56 frame movies. |
| EM imaging optics | Energyfilter name : GIF Bioquantum / Energyfilter slit width: 20 eV : GIF Bioquantum / Energyfilter slit width: 20 eV |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
CTF correction | Details: Patch CTF estimation (multi) from cryoSPARC. / Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 605848 Details: Particle picking was performed using the template picker from cryoSPARC with templates generated from an ab initio model from the same dataset. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
3D reconstruction | Resolution: 5.14 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 166751 / Num. of class averages: 1 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: FLEXIBLE FIT / Space: REAL / Target criteria: Correlation Coefficient | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | Cor.coef. Fo:Fc: 0.975 / WRfactor Rwork: 0.562 / SU B: 3.157 / SU ML: 0.055 / Average fsc overall: 0.0971 / Average fsc work: 0.0971 / Cross valid method: NONE / ESU R: 0.304 Details: Hydrogens have been added in their riding positions
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: BABINET MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 219.884 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: ELECTRON MICROSCOPY / Num. reflection Rfree: 0 / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller



















 PDBj
PDBj





























