-Search query
-Search result
Showing 1 - 50 of 104 items for (author: wang & yx)

EMDB-35618: 
Cryo-EM structure of porcine bc1 complex in isolated state

EMDB-37375: 
Allosteric regulation of nitrate transporter NRT via the signalling protein PII

EMDB-37644: 
Cryo-EM structure of cyanobacterial nitrate/nitrite transporter NrtBCD in complex with signalling protein PII

EMDB-37645: 
Cryo-EM structure of cyanobacterial nitrate/nitrite transporter NrtBCD in complex with nitrate

EMDB-35461: 
Protomer 1 and 2 of the asymmetry trimer of the Cul2-Rbx1-EloBC-FEM1B ubiquitin ligase complex

EMDB-36182: 
An asymmetry dimer of the Cul2-Rbx1-EloBC-FEM1B ubiquitin ligase complexed with BEX2

EMDB-36183: 
Cryo-EM structure of neddylated Cul2-Rbx1-EloBC-FEM1B complexed with FNIP1-FLCN

EMDB-37795: 
Cry-EM structure of cannabinoid receptor-arrestin 2 complex
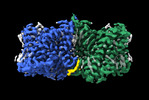
EMDB-36448: 
Structure of AE2 in complex with PIP2

EMDB-36449: 
Structure of R932A/K1147A/H1148A mutant AE2

EMDB-33990: 
Cryo-EM structure of EBV gHgL-gp42 in complex with mAbs 3E8 and 5E3 (localized refinement)
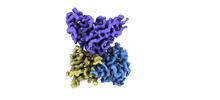
EMDB-33992: 
Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 10E4 (localized refinement)
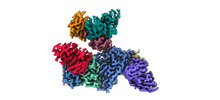
EMDB-33993: 
Cryo-EM density map of EBV gHgL-gp42 in complex with four mAbs 5E3, 3E8, 6H2 and 10E4
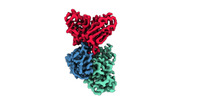
EMDB-33994: 
Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 6H2 (localized refinement)

EMDB-36691: 
Cryo-EM structure of human ABC transporter ABCC2

EMDB-36709: 
Cryo-EM structure of bilirubin ditaurate (BDT) bound human ABC transporter ABCC2

EMDB-36713: 
Cryo-EM structure of human ABC transporter ABCC2 under active turnover condition

EMDB-36719: 
Cryo-EM structure of human ABC transporter ABCC2 in apo' state

EMDB-36720: 
Cryo-EM structure of human ABC transporter ABCC2 in apo" state

EMDB-36342: 
Cryo-EM structure of the beta2AR-mBRIL/1b3 Fab/Glue complex with a partial agonist

EMDB-36360: 
cryo-EM structure of the beta2-AR-mBRIL/1b3 Fab/Glue complex with a full agonist

EMDB-36361: 
Cryo-EM structure of the beta2AR-mBRIL/1b3 Fab/Glue complex with an antagonist

EMDB-40262: 
Adenosylcobalamin-bound riboswitch dimer, form 1

EMDB-40263: 
Adenosylcobalamin-bound riboswitch dimer, form 2

EMDB-40264: 
Adenosylcobalamin-bound riboswitch dimer, form 3

EMDB-40265: 
Adenosylcobalamin-bound riboswitch dimer, form 4

EMDB-40266: 
apo form of adenosylcobalamin riboswitch dimer

EMDB-29903: 
Structure of LARP7 protein p65-telomerase RNA complex in telomerase

EMDB-35043: 
Cryo-EM structure of ABC transporter ABCC3

EMDB-35049: 
Cryo-EM structure of beta-estradiol 17-(beta-D-glucuronide)-bound human ABC transporter ABCC3 in nanodiscs

EMDB-35050: 
Cryo-EM structure of dehydroepiandrosterone sulfate-bound human ABC transporter ABCC3 in nanodiscs

EMDB-32356: 
Computational design of a potent Epstein-Barr virus fusion protein gB nanoparticle vaccine

EMDB-33310: 
Cryo-EM structure of CopC-CaM-caspase-3 with NAD+

EMDB-33311: 
Cryo-EM structure of CopC-CaM-caspase-3 with ADPR

EMDB-33312: 
Cryo-EM structure of CopC-CaM-caspase-3 with ADPR-deacylization

EMDB-26313: 
C6-guano bound Mu Opioid Receptor-Gi Protein Complex

EMDB-32088: 
Computational design of a potent Epstein-Barr virus fusion protein gB nanoparticle vaccine
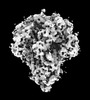
EMDB-32329: 
Cryo-EM map of PEDV (Pintung 52) S protein with all three protomers in the D0-down conformation determined in situ on intact viral particles.

EMDB-32332: 
Subtomogram averaging of PEDV (Pintung 52) S protein with all three protomers in the D0-down conformation determined in situ on intact viral particles.

EMDB-32333: 
Subtomogram averaging of PEDV (Pintung 52) S protein with one protomer in the D0-up conformation and two protomers in the D0-down conformation, determined in situ on intact viral particles

EMDB-32337: 
Subtomogram averaging of PEDV (Pintung 52) S protein with two protomers in the D0-up conformation and one protomer in the D0-down conformation, determined in situ on intact viral particles.

EMDB-32338: 
Cryo-EM map of PEDV S protein with one protomer in the D0-up conformation while the other two in the D0-down conformation
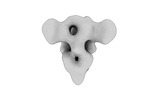
EMDB-32339: 
Subtomogram averaging of PEDV (Pintung 52) S protein with all three protomers in the D0-up conformation determined in situ on intact viral particles.

EMDB-32340: 
Subtomogram averaging of PEDV (Pintung 52) S protein in the postfusion form determined in situ on intact viral particles.

EMDB-33646: 
Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein with three D0-up

EMDB-33647: 
Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein one D0-down and two D0-up

EMDB-33648: 
Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-close conformation
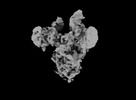
EMDB-33649: 
Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-open conformation

EMDB-33700: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S protein with three D0-down
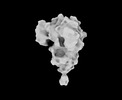
EMDB-33701: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S protein one D0-up and two D0-down
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
