-Search query
-Search result
Showing 1 - 50 of 1,084 items for (author: hara & m)

EMDB-38215: 
Human GPR34 -Gi complex bound to S3E-LysoPS

EMDB-38217: 
Human GPR34 -Gi complex bound to S3E-LysoPS, receptor focused

PDB-8xbe: 
Human GPR34 -Gi complex bound to S3E-LysoPS

PDB-8xbg: 
Human GPR34 -Gi complex bound to S3E-LysoPS, receptor focused

EMDB-50025: 
Cryo-EM structure of the Pseudomonas aeruginosa PAO1 Type IV pilus

PDB-9ewx: 
Cryo-EM structure of the Pseudomonas aeruginosa PAO1 Type IV pilus

EMDB-16489: 
In situ structure of the Nitrosopumilus maritimus S-layer - Six-fold symmetry (C6)

EMDB-16492: 
In situ structure of the Nitrosopumilus maritimus S-layer - Composite map between C2 and C6

PDB-8c8o: 
In situ structure of the Nitrosopumilus maritimus S-layer - Six-fold symmetry (C6)

PDB-8c8r: 
In situ structure of the Nitrosopumilus maritimus S-layer - Composite map between C2 and C6

EMDB-18334: 
Cryo-EM structure of the inward-facing FLVCR1

EMDB-18335: 
Cryo-EM structure of the inward-facing choline-bound FLVCR1

EMDB-18336: 
Cryo-EM structure of the inward-facing FLVCR2
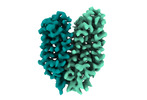
EMDB-18337: 
Cryo-EM structure of the outward-facing FLVCR2

EMDB-18339: 
Cryo-EM structure of the inward-facing choline-bound FLVCR2

EMDB-19009: 
Cryo-EM structure of the inward-facing ethanolamine-bound FLVCR1

PDB-8qcs: 
Cryo-EM structure of the inward-facing FLVCR1

PDB-8qct: 
Cryo-EM structure of the inward-facing choline-bound FLVCR1

PDB-8qcx: 
Cryo-EM structure of the inward-facing FLVCR2

PDB-8qcy: 
Cryo-EM structure of the outward-facing FLVCR2

PDB-8qd0: 
Cryo-EM structure of the inward-facing choline-bound FLVCR2

PDB-8r8t: 
Cryo-EM structure of the inward-facing ethanolamine-bound FLVCR1
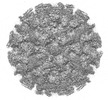
EMDB-36223: 
Cryo-EM structure of the GI.4 Chiba VLP complexed with the CV-1A1 Fv-clasp

PDB-8jg5: 
Cryo-EM structure of the GI.4 Chiba VLP complexed with the CV-1A1 Fv-clasp

EMDB-35163: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 5.5

EMDB-35164: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in closed state
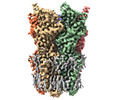
EMDB-36339: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 2.5
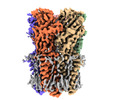
EMDB-37446: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in intermediate state
EMDB-37447: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in open state

PDB-8i47: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 5.5

PDB-8i48: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in closed state

PDB-8jj3: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 2.5

PDB-8wcq: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in intermediate state

PDB-8wcr: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in open state

EMDB-16482: 
In vitro structure of the Nitrosopumilus maritimus S-layer - Six-fold symmetry (C6)

EMDB-16483: 
In vitro structure of the Nitrosopumilus maritimus S-layer - Two-fold symmetry (C2)

EMDB-16484: 
In vitro structure of the Nitrosopumilus maritimus S-layer - Composite map between two and six-fold symmetrised

EMDB-16487: 
In situ structure of the Nitrosopumilus maritimus S-layer - Two-fold symmetry (C2)

PDB-8c8k: 
In vitro structure of the Nitrosopumilus maritimus S-layer - Six-fold symmetry (C6)

PDB-8c8l: 
In vitro structure of the Nitrosopumilus maritimus S-layer - Two-fold symmetry (C2)

PDB-8c8m: 
In vitro structure of the Nitrosopumilus maritimus S-layer - Composite map between two and six-fold symmetrised

PDB-8c8n: 
In situ structure of the Nitrosopumilus maritimus S-layer - Two-fold symmetry (C2)

EMDB-35161: 
Cryo-EM structure of nanodisc (asolectin) reconstituted GLIC at pH 7.5

EMDB-35162: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 7.5

PDB-8i41: 
Cryo-EM structure of nanodisc (asolectin) reconstituted GLIC at pH 7.5

PDB-8i42: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 7.5

EMDB-39463: 
2.6A b-Galactosidase Structure Solved Using an Indirect Scintillator-Coupled CMOS Detector at 300 kV

EMDB-39481: 
2.08A Apoferritin Structure Solved Using an Indirect Scintillator-Coupled CMOS Detector at 300 kV

EMDB-16486: 
In vitro Nitrosopumilus maritimus S-layer with NH4Cl

EMDB-40411: 
PHF Tau from Down Syndrome
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
