-Search query
-Search result
Showing 1 - 50 of 54 items for (author: cash & j)

EMDB-41621: 
Full-length P-Rex1 in complex with inositol 1,3,4,5-tetrakisphosphate (IP4)

PDB-8tua: 
Full-length P-Rex1 in complex with inositol 1,3,4,5-tetrakisphosphate (IP4)

EMDB-40650: 
Phosphoinositide phosphate 3 kinase gamma

EMDB-40651: 
Phosphoinositide phosphate 3 kinase gamma bound with ATP

EMDB-40652: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP

EMDB-40653: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP and Gbetagamma

EMDB-40654: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP and two Gbetagamma subunits in State 1

EMDB-40655: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP and two Gbetagamma subunits in State 2

EMDB-41830: 
Lipidated recombinant apolipoprotein E4

EMDB-41831: 
Gradient-fixed lipidated recombinant apolipoprotein E4

EMDB-25153: 
Human Trio residues 1284-1959 in complex with Rac1

PDB-7sj4: 
Human Trio residues 1284-1959 in complex with Rac1

EMDB-21692: 
PKA RIIbeta holoenzyme with DnaJB1-PKAc fusion in fibrolamellar hepatoceullar carcinoma

EMDB-21693: 
PKA RIIbeta holoenzyme with DnaJB1-PKAc fusion in fibrolamellar hepatoceullar carcinoma

PDB-6wjf: 
PKA RIIbeta holoenzyme with DnaJB1-PKAc fusion in fibrolamellar hepatoceullar carcinoma

PDB-6wjg: 
PKA RIIbeta holoenzyme with DnaJB1-PKAc fusion in fibrolamellar hepatoceullar carcinoma

EMDB-22754: 
Aldolase, rabbit muscle (no beam-tilt refinement)

EMDB-22755: 
Aldolase, rabbit muscle (beam-tilt refinement x1)

EMDB-22756: 
Aldolase, rabbit muscle (beam-tilt refinement x2)

EMDB-22757: 
Aldolase, rabbit muscle (beam-tilt refinement x3)

EMDB-22758: 
Aldolase, rabbit muscle (beam-tilt refinement x4)

PDB-7k9l: 
Aldolase, rabbit muscle (no beam-tilt refinement)

PDB-7k9x: 
Aldolase, rabbit muscle (beam-tilt refinement x1)

PDB-7ka2: 
Aldolase, rabbit muscle (beam-tilt refinement x2)

PDB-7ka3: 
Aldolase, rabbit muscle (beam-tilt refinement x3)

PDB-7ka4: 
Aldolase, rabbit muscle (beam-tilt refinement x4)

EMDB-21490: 
Influenza hemagglutinin trimer preprocessed with automatic workflow

EMDB-21491: 
Thermoplasma acidophilum 20S proteasome cryo-EM structure using automatic preprocessing workflow

EMDB-21492: 
Rabbit muscle aldolase cryo-EM structure using automatic preprocessing workflow

EMDB-20308: 
Single Particle Reconstruction of Phosphatidylinositol (3,4,5) trisphosphate-dependent Rac exchanger 1 bound to G protein beta gamma subunits

PDB-6pcv: 
Single Particle Reconstruction of Phosphatidylinositol (3,4,5) trisphosphate-dependent Rac exchanger 1 bound to G protein beta gamma subunits

EMDB-6442: 
CryoEM structure of endogenously assembled Tetrahymena telomerase holoenzyme at 9.4 Angstrom resolution

EMDB-6443: 
CryoEM structure of endogenously assembled Tetrahymena telomerase holoenzyme at 8.9 Angstrom resolution
EMDB-5807: 
Negative-stain electron microscopy reconstruction of Tetrahymena telomerase (Teb1-f, conformation II)
EMDB-5808: 
Negative-stain electron microscopy reconstruction of Tetrahymena telomerase (Teb1-f, conformation III)
EMDB-5809: 
Negative-stain electron microscopy reconstruction of Tetrahymena telomerase (Teb1-f, conformation IV)
EMDB-5810: 
Negative-stain electron microscopy reconstruction of Tetrahymena telomerase (Teb1-f, conformation V)
EMDB-5811: 
Negative-stain electron microscopy reconstruction of Tetrahymena telomerase (Teb1-f, conformation VI)

EMDB-5812: 
Negative-stain electron microscopy reconstruction of Tetrahymena telomerase (Teb1-f, conformation VII)
EMDB-5813: 
Negative-stain electron microscopy reconstruction of Tetrahymena telomerase (Teb1-f, conformation VIII)
EMDB-5814: 
Negative-stain electron microscopy reconstruction of Tetrahymena telomerase (Teb1-f, conformation IX)
EMDB-5815: 
Negative-stain electron microscopy reconstruction of Tetrahymena telomerase (Teb1-f, conformation X)
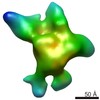
EMDB-5816: 
Negative-stain electron microscopy reconstruction of Tetrahymena telomerase (TERT-f, stable conformation)

EMDB-5817: 
Negative-stain electron microscopy reconstruction of Tetrahymena telomerase (TERT-f, subcomplex lacking p65 N-terminus)

EMDB-5818: 
Negative-stain electron microscopy reconstruction of Tetrahymena telomerase (TERT-f, subcomplex lacking Teb1)
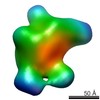
EMDB-5819: 
Negative-stain electron microscopy reconstruction of Tetrahymena telomerase (TERT-f, subcomplex lacking p75-p19-p45)
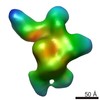
EMDB-5820: 
Negative-stain electron microscopy reconstruction of Tetrahymena telomerase (f-Teb1C, stable conformation)
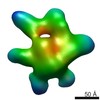
EMDB-5821: 
Negative-stain electron microscopy reconstruction of Tetrahymena telomerase (TERT-f, affinity labeled by anti-FLAG Fab)

EMDB-5822: 
Negative-stain electron microscopy reconstruction of Tetrahymena telomerase (Teb1-f, affinity labeled by anti-FLAG Fab)
EMDB-5823: 
Negative-stain electron microscopy reconstruction of Tetrahymena telomerase (f-p50, affinity labeled by two copies of anti-FLAG Fab)
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
