-Search query
-Search result
Showing 1 - 50 of 123 items for (author: wu & ym)

EMDB-41363: 
Cryo-EM structure of DDB1deltaB-DDA1-DCAF5
Method: single particle / : Yue H, Hunkeler M, Roy Burman SS, Fischer ES

PDB-8tl6: 
Cryo-EM structure of DDB1deltaB-DDA1-DCAF5
Method: single particle / : Yue H, Hunkeler M, Roy Burman SS, Fischer ES

EMDB-16626: 
Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium, 3'-phosphoadenosine-5'-phosphosulfate, and nanobody nAb7 (original map)
Method: single particle / : Mycroft-West CJ, Wu L

EMDB-16627: 
Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium, 3'-phosphoadenosine-5'-phosphosulfate, and nanobody nAb7 (locally refined map of N-terminal and deacetylase domains)
Method: single particle / : Mycroft-West CJ, Wu L

EMDB-16629: 
Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium, 3'-phosphoadenosine-5'-phosphosulfate, and nanobody nAb7 (locally refined map of deacetylase and sulfotransferase domains)
Method: single particle / : Mycroft-West CJ, Wu L

EMDB-16661: 
Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium, 3'-phosphoadenosine-5'-phosphosulfate, and nanobody nAb13 (original map)
Method: single particle / : Mycroft-West CJ, Wu L

EMDB-16662: 
Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium, 3'-phosphoadenosine-5'-phosphosulfate, and nanobody nAb13 (locally refined map of N-terminal and deacetylase domains)
Method: single particle / : Mycroft-West CJ, Wu L

EMDB-16663: 
Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium, 3'-phosphoadenosine-5'-phosphosulfate, and nanobody nAb13 (locally refined map of deacetylase and sulfotransferase domains)
Method: single particle / : Mycroft-West CJ, Wu L

EMDB-16664: 
Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium, 3'-phosphoadenosine-5'-phosphosulfate and nanobody nAb13 (composite map and model).
Method: single particle / : Mycroft-West CJ, Wu L

PDB-8chs: 
Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium, 3'-phosphoadenosine-5'-phosphosulfate and nanobody nAb13 (composite map and model).
Method: single particle / : Mycroft-West CJ, Wu L

EMDB-16564: 
Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium and 3'-phosphoadenosine-5'-phosphosulfate
Method: single particle / : Mycroft-West CJ, Wu L

EMDB-16565: 
Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium, 3'-phosphoadenosine-5'-phosphosulfate, and nanobody nAb7 (composite map and model)
Method: single particle / : Mycroft-West CJ, Wu L

PDB-8ccy: 
Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium and 3'-phosphoadenosine-5'-phosphosulfate
Method: single particle / : Mycroft-West CJ, Wu L

PDB-8cd0: 
Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium, 3'-phosphoadenosine-5'-phosphosulfate, and nanobody nAb7 (composite map and model)
Method: single particle / : Mycroft-West CJ, Wu L

EMDB-36076: 
Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class2
Method: single particle / : Yang GH, Zhang YM, Zhou JQ, Jia YT, Xu X, Fu P, Wu HY

EMDB-36077: 
Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class1
Method: single particle / : Yang GH, Zhang YM, Zhou JQ, Jia YT, Xu X, Fu P, Wu HY

EMDB-34490: 
The cryo-EM structure of nuclear transport receptor Kap114p complex with yeast TATA-box binding protein
Method: single particle / : Hsia KC, Liao CC, Wang CH, Wu YM

EMDB-34030: 
Cryo-EM map of a dimeric form of Ecoli Malate Synthase G (MSG)
Method: single particle / : Wu KP, Wu YM, Lu YC

EMDB-34029: 
2.9-angstrom cryo-EM structure of Ecoli malate synthase G
Method: single particle / : Wu KP, Wu YM, Lu YC

EMDB-33241: 
Cryo-EM Structure of Human Niacin Receptor HCA2-Gi protein complex
Method: single particle / : Yang Y, Kang HJ, Gao RG, Wang JJ, Han GW, DiBerto JF, Wu LJ, Tong JH, Qu L, Wu YR, Pileski R, Li XM, Zhang XC, Zhao SW, Kenakin T, Wang Q, Stevens RC, Peng W, Roth BL, Rao ZH, Liu ZJ

PDB-7xk2: 
Cryo-EM Structure of Human Niacin Receptor HCA2-Gi protein complex
Method: single particle / : Yang Y, Kang HJ, Gao RG, Wang JJ, Han GW, DiBerto JF, Wu LJ, Tong JH, Qu L, Wu YR, Pileski R, Li XM, Zhang XC, Zhao SW, Kenakin T, Wang Q, Stevens RC, Peng W, Roth BL, Rao ZH, Liu ZJ

EMDB-34806: 
SARS-CoV-2 Delta Spike in complex with FP-12A
Method: single particle / : Chen X, Wu YM

EMDB-34807: 
SARS-CoV-2 Delta Spike in complex with IS-9A
Method: single particle / : Mohapatra A, Wu YM

EMDB-34808: 
SARS-CoV-2 Omicron BA.1 Spike in complex with IY-2A
Method: single particle / : Chen X, Mohapatra A, Wu YM
EMDB-32329: 
Cryo-EM map of PEDV (Pintung 52) S protein with all three protomers in the D0-down conformation determined in situ on intact viral particles.
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-32332: 
Subtomogram averaging of PEDV (Pintung 52) S protein with all three protomers in the D0-down conformation determined in situ on intact viral particles.
Method: subtomogram averaging / : Hsu STD, Draczkowski P, Wang YS, Huang CY

EMDB-32333: 
Subtomogram averaging of PEDV (Pintung 52) S protein with one protomer in the D0-up conformation and two protomers in the D0-down conformation, determined in situ on intact viral particles
Method: subtomogram averaging / : Hsu STD, Draczkowski P, Wang YS, Huang CY

EMDB-32337: 
Subtomogram averaging of PEDV (Pintung 52) S protein with two protomers in the D0-up conformation and one protomer in the D0-down conformation, determined in situ on intact viral particles.
Method: subtomogram averaging / : Hsu STD, Draczkowski P, Wang YS, Huang CY

EMDB-32338: 
Cryo-EM map of PEDV S protein with one protomer in the D0-up conformation while the other two in the D0-down conformation
Method: single particle / : Hsu STD, Draczkowski P, Wang YS
EMDB-32339: 
Subtomogram averaging of PEDV (Pintung 52) S protein with all three protomers in the D0-up conformation determined in situ on intact viral particles.
Method: subtomogram averaging / : Hsu STD, Draczkowski P, Wang YS, Huang CY

EMDB-32340: 
Subtomogram averaging of PEDV (Pintung 52) S protein in the postfusion form determined in situ on intact viral particles.
Method: subtomogram averaging / : Hsu STD, Draczkowski P, Wang YS, Huang CY

EMDB-33646: 
Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein with three D0-up
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-33647: 
Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein one D0-down and two D0-up
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-33648: 
Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-close conformation
Method: single particle / : Hsu STD, Draczkowski P, Wang YS
EMDB-33649: 
Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-open conformation
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-33700: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S protein with three D0-down
Method: single particle / : Hsu STD, Draczkowski P, Wang YS
EMDB-33701: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S protein one D0-up and two D0-down
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-33702: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S protein with three D0-up
Method: single particle / : Hsu STD, Draczkowski P, Wang YS
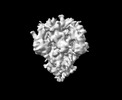
EMDB-33703: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S T326I with three D0-down
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-33704: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S T326I one D0-up and two D0-down
Method: single particle / : Hsu STD, Draczkowski P, Wang YS
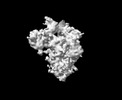
EMDB-33705: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S T326I one D0-down and two D0-up
Method: single particle / : Hsu STD, Draczkowski P, Wang YS
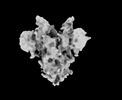
EMDB-33706: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S T326I with three D0-up
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-32243: 
Cryo-EM structure of a dimeric GPCR-Gi complex with small molecule
Method: single particle / : Yue Y, Liu LE, Wu LJ, Xu F, Hanson M

EMDB-32244: 
Cryo-EM structure of a monomeric GPCR-Gi complex with small molecule
Method: single particle / : Xu F, Yue Y, Liu LE, Wu LJ, Hanson M

EMDB-32245: 
Cryo-EM structure of a dimeric GPCR-Gi complex with peptide
Method: single particle / : Xu F, Yue Y, Wu LJ, Liu LE, Hanson M

EMDB-32246: 
Cryo-EM structure of a monomeric GPCR-Gi complex with peptide
Method: single particle / : Xu F, Yue Y, Liu LE, Wu LJ, Hanson M

EMDB-32247: 
Cryo-EM structure of a GPCR-Gi complex with peptide
Method: single particle / : Xu F, Yue Y, Liu LE, Wu LJ, Hanson M

PDB-7w0l: 
Cryo-EM structure of a dimeric GPCR-Gi complex with small molecule
Method: single particle / : Yue Y, Liu LE, Wu LJ, Xu F, Hanson M

PDB-7w0m: 
Cryo-EM structure of a monomeric GPCR-Gi complex with small molecule
Method: single particle / : Xu F, Yue Y, Liu LE, Wu LJ, Hanson M

PDB-7w0n: 
Cryo-EM structure of a dimeric GPCR-Gi complex with peptide
Method: single particle / : Xu F, Yue Y, Wu LJ, Liu LE, Hanson M
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
