-Search query
-Search result
Showing 1 - 50 of 663 items for (author: san & martin & c)
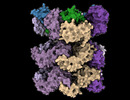
EMDB-19177: 
Structure of the 55LCC ATPase complex
Method: single particle / : Foglizzo M, Degtjarik O, Zeqiraj E

PDB-8rhn: 
Structure of the 55LCC ATPase complex
Method: single particle / : Foglizzo M, Degtjarik O, Zeqiraj E

EMDB-19477: 
Saccharomyces cerevisiae FAS type I
Method: single particle / : Mann D, Grininger M, Ludig D, Sachse C

EMDB-19489: 
Tobacco mosaic virus from scanning transmission electron microscopy at CSA=2.0 mrad
Method: helical / : Mann D, Filopoulou A, Sachse C

EMDB-43658: 
SARS-CoV-2 S (C.37 Lambda variant) plus S309, S2L20, and S2X303 Fabs
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-43659: 
SARS-CoV-2 S NTD (C.37 Lambda variant) plus S2L20 and S2X303 Fabs, local refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-43660: 
SARS-CoV-2 S RBD (C.37 Lambda variant) plus S309 Fab, local refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-8vye: 
SARS-CoV-2 S (C.37 Lambda variant) plus S309, S2L20, and S2X303 Fabs
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-8vyf: 
SARS-CoV-2 S NTD (C.37 Lambda variant) plus S2L20 and S2X303 Fabs, local refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-8vyg: 
SARS-CoV-2 S RBD (C.37 Lambda variant) plus S309 Fab, local refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)
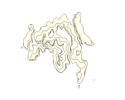
EMDB-19184: 
Late alpha-Synuclein fibril structure from liquid-liquid phase separations.
Method: helical / : De Simone A, Barritt JD, Chen S, Cascella R, Cecchi C, Bigi A, Jarvis JA, Chiti F, Dobson CM, Fusco G

PDB-8ri9: 
Late alpha-Synuclein fibril structure from liquid-liquid phase separations.
Method: helical / : De Simone A, Barritt JD, Chen S, Cascella R, Cecchi C, Bigi A, Jarvis JA, Chiti F, Dobson CM, Fusco G

EMDB-15474: 
In situ cryo-electron tomogram of an intact primary neuronal process
Method: electron tomography / : Chakraborty S, Martinez-Sanchez A, Baumeister W, Mahamid J

EMDB-15454: 
Subtomogram average of bound microtubule inner protein 2
Method: subtomogram averaging / : Chakraborty S, Martinez-Sanchez A, Baumeister W, Mahamid J

EMDB-15457: 
Subtomogram average of floating microtubule inner protein 5
Method: subtomogram averaging / : Chakraborty S, Mahamid J, Martinez-Sanchez A, Baumeister W

EMDB-15463: 
Subtomogram average of floating microtubule inner protein 12
Method: subtomogram averaging / : Chakraborty S, Mahamid J, Martinez-Sanchez A, Baumeister W

EMDB-15456: 
In situ subtomogram average of bound microtubule inner protein 4 (bMIP4)
Method: subtomogram averaging / : Chakraborty S, Martinez-Sanchez A, Baumeister W, Mahamid J

EMDB-15459: 
In situ subtomogram average of floating microtubule inner protein 7
Method: subtomogram averaging / : Chakraborty S, Mahamid J, Martinez-Sanchez A, Baumeister W

EMDB-15464: 
In situ subtomogram average of floating microtubule inner protein 13 (fMIP13)
Method: subtomogram averaging / : Chakraborty S, Mahamid J, Martinez-Sanchez A, Baumeister W

EMDB-15458: 
Subtomogram average of floating microtubule inner protein 6
Method: subtomogram averaging / : Chakraborty S, Mahamid J, Martinez-Sanchez A, Baumeister W

EMDB-15461: 
Subtomogram average of floating microtubule inner protein 10
Method: subtomogram averaging / : Chakraborty S, Mahamid J, Martinez-Sanchez A, Baumeister W

EMDB-15472: 
In situ subtomogram average of floating microtubule inner protein 8
Method: subtomogram averaging / : Chakraborty S, Mahamid J, Martinez-Sanchez A, Baumeister W

EMDB-15469: 
Subtomogram average of murine 80S ribosome
Method: subtomogram averaging / : Chakraborty S, Mahamid J, Martinez-Sanchez A, Baumeister W

EMDB-15460: 
Subtomogram average of floating microtubule inner protein 9
Method: subtomogram averaging / : Chakraborty S, Mahamid J, Martinez-Sanchez A, Baumeister W

EMDB-15455: 
Subtomogram average of bound microtubule inner protein 3
Method: subtomogram averaging / : Chakraborty S, Martinez-Sanchez A, Baumeister W, Mahamid J

EMDB-15444: 
Subtomogram average of floating microtubule inner protein 4
Method: subtomogram averaging / : Chakraborty S, Martinez-Sanchez A, Baumeister W, Mahamid J

EMDB-15467: 
In situ subtomogram average of floating microtubule inner protein 15 (fMIP15)
Method: subtomogram averaging / : Chakraborty S, Mahamid J, Martinez-Sanchez A, Baumeister W

EMDB-15470: 
In situ subtomogram average of primary neuronal microtubules
Method: subtomogram averaging / : Chakraborty S, Mahamid J, Martinez-Sanchez A, Baumeister W

EMDB-15466: 
In situ subtomogram average of floating microtubule inner protein 14 (fMIP14)
Method: subtomogram averaging / : Chakraborty S, Mahamid J, Martinez-Sanchez A, Baumeister W

EMDB-15453: 
In situ subtomogram average of bound microtubule inner protein 1 (bMIP1)
Method: subtomogram averaging / : Chakraborty S, Martinez-Sanchez A, Baumeister W, Mahamid J

EMDB-15468: 
In situ subtomogram average of floating microtubule inner protein 16 (fMIP16)
Method: subtomogram averaging / : Chakraborty S, Mahamid J, Martinez-Sanchez A, Baumeister W

EMDB-15462: 
Subtomogram average of floating microtubule inner protein 11
Method: subtomogram averaging / : Chakraborty S, Mahamid J, Martinez-Sanchez A, Baumeister W

EMDB-15440: 
In situ cryo-electron tomogram of an intact murine P19 cellular process.
Method: electron tomography / : Chakraborty S, Martinez-Sanchez A, Baumeister W, Mahamid J

EMDB-15437: 
In situ cryo-electron tomogram of an intact human induced pluripotent stem cell derived neuronal process
Method: electron tomography / : Chakraborty S, Martinez-Sanchez A, Baumeister W, Mahamid J

EMDB-15438: 
In situ cryo-electron tomogram of an intact differentiated P19 neuronal process
Method: electron tomography / : Chakraborty S, Martinez-Sanchez A, Baumeister W, Mahamid J

EMDB-15442: 
In situ cryo-electron tomogram of an intact pluripotent P19 cellular process after nocodazole treatment and washout.
Method: electron tomography / : Chakraborty S, Martinez-Sanchez A, Baumeister W, Mahamid J

EMDB-15443: 
In situ cryo-electron tomogram of a Taxol treated intact pluripotent P19 cell.
Method: electron tomography / : Chakraborty S, Martinez-Sanchez A, Baumeister W, Mahamid J

EMDB-15439: 
Subtomogram average of floating microtubule inner protein 2 (fMIP2)
Method: subtomogram averaging / : Chakraborty S, Martinez-Sanchez A, Baumeister W, Mahamid J

EMDB-15436: 
In situ subtomogram average of floating microtubule inner protein 3 (fMIP3)
Method: subtomogram averaging / : Chakraborty S, Martinez-Sanchez A, Baumeister W, Mahamid J

EMDB-15441: 
In situ subtomogram average of floating microtubule inner protein 1 (fMIP1)
Method: subtomogram averaging / : Chakraborty S, Martinez-Sanchez A, Baumeister W, Mahamid J

EMDB-15411: 
Single particle structure of Atg18-WT
Method: single particle / : Mann D, Fromm S, Martinez-Sanchez A, Gopaldass N, Mayer A, Sachse C

PDB-8afx: 
Single particle structure of Atg18-WT
Method: single particle / : Mann D, Fromm S, Martinez-Sanchez A, Gopaldass N, Mayer A, Sachse C

EMDB-40856: 
Single particle reconstruction of the human LINE-1 ORF2p without substrate (apo)
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
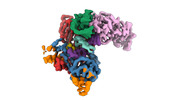
EMDB-40858: 
Structure of LINE-1 ORF2p with template:primer hybrid
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
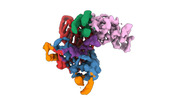
EMDB-40859: 
Structure of LINE-1 ORF2p with an oligo(A) template
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

PDB-8sxt: 
Structure of LINE-1 ORF2p with template:primer hybrid
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

PDB-8sxu: 
Structure of LINE-1 ORF2p with an oligo(A) template
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

EMDB-17402: 
Uncharacterized Q8U0N8 protein from Pyrococcus furiosus
Method: single particle / : Pacesa M, Correia BE, Levy ED

EMDB-18415: 
Cysteine tRNA ligase homodimer
Method: single particle / : Pacesa M, Correia BE, Levy ED

PDB-8p49: 
Uncharacterized Q8U0N8 protein from Pyrococcus furiosus
Method: single particle / : Pacesa M, Correia BE, Levy ED
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
