+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Uncharacterized Q8U0N8 protein from Pyrococcus furiosus | |||||||||
 Map data Map data | Q8U0N8 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | uncharacterized /  hexamer / hexamer /  pyrococcus / pore / UNKNOWN FUNCTION pyrococcus / pore / UNKNOWN FUNCTION | |||||||||
| Function / homology | Uncharacterized protein Function and homology information Function and homology information | |||||||||
| Biological species |    Pyrococcus furiosus DSM 3638 (archaea) Pyrococcus furiosus DSM 3638 (archaea) | |||||||||
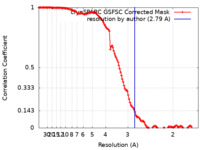
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.79 Å cryo EM / Resolution: 2.79 Å | |||||||||
 Authors Authors | Pacesa M / Correia BE / Levy ED | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2024 Journal: Cell / Year: 2024Title: An atlas of protein homo-oligomerization across domains of life. Authors: Hugo Schweke / Martin Pacesa / Tal Levin / Casper A Goverde / Prasun Kumar / Yoan Duhoo / Lars J Dornfeld / Benjamin Dubreuil / Sandrine Georgeon / Sergey Ovchinnikov / Derek N Woolfson / ...Authors: Hugo Schweke / Martin Pacesa / Tal Levin / Casper A Goverde / Prasun Kumar / Yoan Duhoo / Lars J Dornfeld / Benjamin Dubreuil / Sandrine Georgeon / Sergey Ovchinnikov / Derek N Woolfson / Bruno E Correia / Sucharita Dey / Emmanuel D Levy /      Abstract: Protein structures are essential to understanding cellular processes in molecular detail. While advances in artificial intelligence revealed the tertiary structure of proteins at scale, their ...Protein structures are essential to understanding cellular processes in molecular detail. While advances in artificial intelligence revealed the tertiary structure of proteins at scale, their quaternary structure remains mostly unknown. We devise a scalable strategy based on AlphaFold2 to predict homo-oligomeric assemblies across four proteomes spanning the tree of life. Our results suggest that approximately 45% of an archaeal proteome and a bacterial proteome and 20% of two eukaryotic proteomes form homomers. Our predictions accurately capture protein homo-oligomerization, recapitulate megadalton complexes, and unveil hundreds of homo-oligomer types, including three confirmed experimentally by structure determination. Integrating these datasets with omics information suggests that a majority of known protein complexes are symmetric. Finally, these datasets provide a structural context for interpreting disease mutations and reveal coiled-coil regions as major enablers of quaternary structure evolution in human. Our strategy is applicable to any organism and provides a comprehensive view of homo-oligomerization in proteomes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17402.map.gz emd_17402.map.gz | 104.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17402-v30.xml emd-17402-v30.xml emd-17402.xml emd-17402.xml | 17.4 KB 17.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17402_fsc.xml emd_17402_fsc.xml | 12.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_17402.png emd_17402.png | 129.5 KB | ||
| Masks |  emd_17402_msk_1.map emd_17402_msk_1.map | 209.3 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17402.cif.gz emd-17402.cif.gz | 5.9 KB | ||
| Others |  emd_17402_half_map_1.map.gz emd_17402_half_map_1.map.gz emd_17402_half_map_2.map.gz emd_17402_half_map_2.map.gz | 194.5 MB 194.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17402 http://ftp.pdbj.org/pub/emdb/structures/EMD-17402 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17402 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17402 | HTTPS FTP |
-Related structure data
| Related structure data |  8p49MC  8q70C  8qhpC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17402.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17402.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Q8U0N8 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
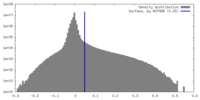
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17402_msk_1.map emd_17402_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
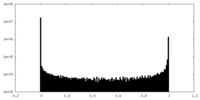
| Density Histograms |
-Half map: half map A
| File | emd_17402_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
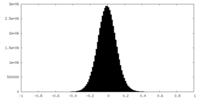
| Density Histograms |
-Half map: half map B
| File | emd_17402_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
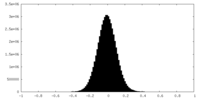
| Density Histograms |
- Sample components
Sample components
-Entire : Hexameric Q8U0N8 uncharacterized protein from Pyrococcus furiosus
| Entire | Name: Hexameric Q8U0N8 uncharacterized protein from Pyrococcus furiosus |
|---|---|
| Components |
|
-Supramolecule #1: Hexameric Q8U0N8 uncharacterized protein from Pyrococcus furiosus
| Supramolecule | Name: Hexameric Q8U0N8 uncharacterized protein from Pyrococcus furiosus type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:    Pyrococcus furiosus DSM 3638 (archaea) Pyrococcus furiosus DSM 3638 (archaea) |
-Macromolecule #1: Q8U0N8 protein
| Macromolecule | Name: Q8U0N8 protein / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Pyrococcus furiosus DSM 3638 (archaea) Pyrococcus furiosus DSM 3638 (archaea) |
| Molecular weight | Theoretical: 46.734129 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MSESINNITK PKERRDFVVM AGMRKDGTID FIKVYALNEK LAIEVLEAFL KENNIHPSDF IVIQRGYEDV KDKKAITTRS EEELSAMLG RLGLRLVSNG VLYTDGIDKL YQITAISREL FESLQKEKRE IFEDVQEKIT FNFSKVDLPE KYVKKLRLLE L MEDTIIFN ...String: MSESINNITK PKERRDFVVM AGMRKDGTID FIKVYALNEK LAIEVLEAFL KENNIHPSDF IVIQRGYEDV KDKKAITTRS EEELSAMLG RLGLRLVSNG VLYTDGIDKL YQITAISREL FESLQKEKRE IFEDVQEKIT FNFSKVDLPE KYVKKLRLLE L MEDTIIFN MAELEIPNLL KAIVEGTVLI PRFLEKEDLI IRIFDEELHE YRGSYFDKVL IKPPIIHWDF YLDSLEDFSF KK VEESIYI APLFLRATGG FLILTEPPED LVKTLLKLKK RGEVRTILEG KRITIPINFT LIVDTRHPER YAGLKFPIRI NLP PLDDET FLKVLETNLG ITPPTEIVRI FPPDYKTFLG VELIKNLFEK LKLTEKGKDE VSLLKEAATI ITGGTPGGSS GGSG HHHHH H UniProtKB: Uncharacterized protein |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 168674 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 96000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 96000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Temperature | Min: 186.0 K / Max: 192.0 K |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Average exposure time: 3.0 sec. / Average electron dose: 60.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
| Output model |  PDB-8p49: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)