-Search query
-Search result
Showing 1 - 50 of 208 items for (author: robert & williams)
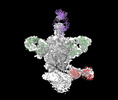
EMDB-44246: 
Cryo-EM structure of HIV-1 JRFL v6 Env in complex with vaccine-elicited, Membrane Proximal External Region (MPER) directed antibody DH1317.4.
Method: single particle / : Acharya P, Parsons R, Janowska K, Williams WB, Alam M, Haynes BF

EMDB-40853: 
CH505 Disulfide Stapled SOSIP Bound to b12 Fab
Method: single particle / : Henderson R
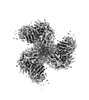
EMDB-40854: 
CH505 Disulfide Stapled SOSIP Bound to CH235.12 Fab
Method: single particle / : Henderson R

PDB-8sxi: 
CH505 Disulfide Stapled SOSIP Bound to b12 Fab
Method: single particle / : Henderson R

PDB-8sxj: 
CH505 Disulfide Stapled SOSIP Bound to CH235.12 Fab
Method: single particle / : Henderson R

EMDB-41810: 
Cryo-EM structure of vaccine-elicited CD4 binding site antibody DH1285 bound to HIV-1 CH505TFchim.6R.SOSIP.664v4.1 Env Local Refinement
Method: single particle / : Thakur B, Stalls VD, Acharya P

EMDB-41820: 
Cryo-EM structure of vaccine-elicited CD4 binding site antibody DH1285 bound to HIV-1 CH505TFchim.6R.SOSIP.664v4.1 Env
Method: single particle / : Thakur B, Stalls VD, Acharya P

EMDB-41823: 
Cryo-EM structure of vaccine-elicited CD4 binding site antibody DH1285 bound to HIV-1 CH505TFchim.6R.SOSIP.664v4.1 Env
Method: single particle / : Thakur B, Stalls VD, Acharya P

EMDB-41838: 
Cryo-EM structure of vaccine-elicited CD4 binding site antibody DH1285 bound to partially open HIV-1 CH505TFchim.6R.SOSIP.664v4.1 Env
Method: single particle / : Thakur B, Stalls VD, Acharya P

PDB-8u1d: 
Cryo-EM structure of vaccine-elicited CD4 binding site antibody DH1285 bound to HIV-1 CH505TFchim.6R.SOSIP.664v4.1 Env Local Refinement
Method: single particle / : Thakur B, Stalls VD, Acharya P
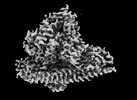
EMDB-41617: 
CryoEM structure of PI3Kalpha
Method: single particle / : Valverde R, Shi H, Holliday M, Sun M

PDB-8tu6: 
CryoEM structure of PI3Kalpha
Method: single particle / : Valverde R, Shi H, Holliday M

EMDB-27808: 
RSV F trimer bound to RSV-199 Fab
Method: single particle / : Wen X, Jardetzky TS

EMDB-27846: 
HMPV F monomer bound to RSV-199 Fab
Method: single particle / : Wen X, Jardetzky TS

EMDB-27990: 
HMPV F complex with 4I3 Fab
Method: single particle / : Wen X, Jardetzky TS

EMDB-27995: 
HMPV F dimer bound to RSV-199 Fab
Method: single particle / : Wen X, Jardetzky TS

PDB-8dzw: 
RSV F trimer bound to RSV-199 Fab
Method: single particle / : Wen X, Jardetzky TS

PDB-8e2u: 
HMPV F monomer bound to RSV-199 Fab
Method: single particle / : Wen X, Jardetzky TS

PDB-8eay: 
HMPV F complex with 4I3 Fab
Method: single particle / : Wen X, Jardetzky TS

PDB-8ebp: 
HMPV F dimer bound to RSV-199 Fab
Method: single particle / : Wen X, Jardetzky TS

EMDB-28755: 
Human tRNA Splicing Endonuclease Complex bound to 2'F-tRNA-Arg
Method: single particle / : Stanley RE, Hayne CK

EMDB-26856: 
Human tRNA Splicing Endonuclease Complex bound to pre-tRNA-ARG
Method: single particle / : Stanley RE, Hayne CK

PDB-7uxa: 
Human tRNA Splicing Endonuclease Complex bound to pre-tRNA-ARG
Method: single particle / : Stanley RE, Hayne CK

EMDB-26005: 
Structure of the Inmazeb cocktail and resistance to escape against Ebola virus
Method: single particle / : Rayaprolu V, Fulton B, Rafique A, Arturo E, Williams D, Hariharan C, Callaway H, Parvate A, Schendel SL, Parekh D, Hui S, Shaffer K, Pascal KE, Wloga E, Giordano S, Copin R, Franklin M, Boytz RM, Donahue C, Davey R, Baum A, Kyratsous CA, Saphire EO

PDB-7tn9: 
Structure of the Inmazeb cocktail and resistance to escape against Ebola virus
Method: single particle / : Rayaprolu V, Fulton B, Rafique A, Arturo E, Williams D, Hariharan C, Callaway H, Parvate A, Schendel SL, Parekh D, Hui S, Shaffer K, Pascal KE, Wloga E, Giordano S, Copin R, Franklin M, Boytz RM, Donahue C, Davey R, Baum A, Kyratsous CA, Saphire EO
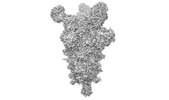
EMDB-26676: 
Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment
Method: single particle / : Zhang J, Luo S, Kreutzberger A, Kirchhausen T, Chen B, Haynes B, Alt F

EMDB-26677: 
Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment (local refinement of the RBD and Fab variable domains)
Method: single particle / : Zhang J, Luo S, Kreutzberger A, Kirchhausen T, Chen B, Haynes B, Alt F

EMDB-26678: 
An antibody from single human VH-rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion
Method: single particle / : Zhang J, Luo S, Kreutzberger A, Kirchhausen T, Chen B, Haynes B, Alt F

EMDB-27043: 
Negative stain electron microscopy single particle reconstruction of monoclonal antibody SP1-77 Fab in complex with SARS-CoV2 2P spike
Method: single particle / : Edwards RJ, Mansouri K
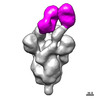
EMDB-27044: 
Negative stain electron microscopy single particle reconstruction of monoclonal antibody VHH7-7-53 Fab in complex with SARS-CoV2 2P spike
Method: single particle / : Edwards RJ, Mansouri K

EMDB-27046: 
Negative stain electron microscopy single particle reconstruction of monoclonal antibody VHH7-5-82 Fab in complex with SARS-CoV2 2P spike
Method: single particle / : Edwards RJ, Mansouri K

PDB-7upw: 
Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment
Method: single particle / : Zhang J, Luo S, Kreutzberger A, Kirchhausen T, Chen B, Haynes B, Alt F

PDB-7upx: 
Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment (local refinement of the RBD and Fab variable domains)
Method: single particle / : Zhang J, Luo S, Kreutzberger A, Kirchhausen T, Chen B, Haynes B, Alt F

PDB-7upy: 
An antibody from single human VH-rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion
Method: single particle / : Zhang J, Luo S, Kreutzberger A, Kirchhausen T, Chen B, Haynes B, Alt F

EMDB-25743: 
Octameric Human Twinkle Helicase Clinical Variant W315L
Method: single particle / : Riccio AA, Bouvette J

EMDB-25744: 
Heptameric Human Twinkle Helicase Clinical Variant W315L
Method: single particle / : Riccio AA, Bouvette J

EMDB-25746: 
Octameric Twinkle Helicase Clinical Variant W315L, local refinement
Method: single particle / : Riccio AA, Bouvette J, Borgnia MJ, Copeland WC

PDB-7t8b: 
Octameric Human Twinkle Helicase Clinical Variant W315L
Method: single particle / : Riccio AA, Bouvette J, Krahn J, Borgnia MJ, Copeland WC

PDB-7t8c: 
Heptameric Human Twinkle Helicase Clinical Variant W315L
Method: single particle / : Riccio AA, Bouvette J, Krahn J, Borgnia MJ, Copeland WC

EMDB-25777: 
Structure of VRK1 C-terminal tail bound to nucleosome core particle
Method: single particle / : Spangler CJ, Budziszewski GR

EMDB-25778: 
Uncrosslinked VRK1-nucleosome complex
Method: single particle / : Spangler CJ, Budziszewski GR, McGinty RK

PDB-7tan: 
Structure of VRK1 C-terminal tail bound to nucleosome core particle
Method: single particle / : Spangler CJ, Budziszewski GR, McGinty RK

EMDB-26433: 
SARS-CoV-2 Omicron-BA.2 3-RBD down Spike Protein Trimer without the P986-P987 stabilizing mutations (S-GSAS-Omicron-BA.2)
Method: single particle / : Stalls V, Acharya P
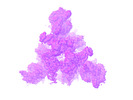
EMDB-26435: 
SARS-CoV-2 Omicron-BA.2 3-RBD down Spike Protein Trimer without the P986-P987 stabilizing mutations (S-GSAS-Omicron-BA.2)
Method: single particle / : Stalls V, Acharya P

EMDB-26436: 
SARS-CoV-2 Omicron-BA.2 3-RBD down Spike Protein Trimer without the P986-P987 stabilizing mutations (S-GSAS-Omicron-BA.2)
Method: single particle / : Stalls V, Acharya P

EMDB-26643: 
SARS-CoV-2 Omicron-BA.2 1.5-RBD up Spike Protein Trimer without the P986-P987 stabilizing mutations (S-GSAS-Omicron-BA.2)
Method: single particle / : Stalls V, Acharya P

EMDB-26644: 
SARS-CoV-2 Omicron-BA.2 1-RBD-up Spike Protein Trimer without the P986-P987 stabilizing mutations (S-GSAS-Omicron-BA.2)
Method: single particle / : Stalls V, Acharya P

EMDB-26647: 
SARS-CoV-2 Omicron-BA.2 1-RBD-up Spike Protein Trimer without the P986-P987 stabilizing mutations (S-GSAS-Omicron-BA.2)
Method: single particle / : Stalls V, Acharya P

PDB-7ub0: 
SARS-CoV-2 Omicron-BA.2 3-RBD down Spike Protein Trimer without the P986-P987 stabilizing mutations (S-GSAS-Omicron-BA.2)
Method: single particle / : Stalls V, Acharya P

PDB-7ub5: 
SARS-CoV-2 Omicron-BA.2 3-RBD down Spike Protein Trimer without the P986-P987 stabilizing mutations (S-GSAS-Omicron-BA.2)
Method: single particle / : Stalls V, Acharya P
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
