[English] 日本語
 Yorodumi
Yorodumi- EMDB-27046: Negative stain electron microscopy single particle reconstruction... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
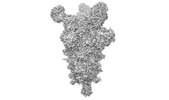
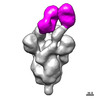
| Title | Negative stain electron microscopy single particle reconstruction of monoclonal antibody VHH7-5-82 Fab in complex with SARS-CoV2 2P spike | ||||||||||||
 Map data Map data | Final sharpened map of negative stain electron microscopy single particle reconstruction of monoclonal antibody VHH7-5-82 in complex with SARS-CoV2 2P spike. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | SARS-CoV2 spike / antibody Fab /  VIRAL PROTEIN VIRAL PROTEIN | ||||||||||||
| Biological species |   Severe acute respiratory syndrome coronavirus / Severe acute respiratory syndrome coronavirus /   Mus musculus (house mouse) Mus musculus (house mouse) | ||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 13.2 Å negative staining / Resolution: 13.2 Å | ||||||||||||
 Authors Authors | Edwards RJ / Mansouri K | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Immunol / Year: 2022 Journal: Sci Immunol / Year: 2022Title: An antibody from single human V-rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion. Authors: Sai Luo / Jun Zhang / Alex J B Kreutzberger / Amanda Eaton / Robert J Edwards / Changbin Jing / Hai-Qiang Dai / Gregory D Sempowski / Kenneth Cronin / Robert Parks / Adam Yongxin Ye / ...Authors: Sai Luo / Jun Zhang / Alex J B Kreutzberger / Amanda Eaton / Robert J Edwards / Changbin Jing / Hai-Qiang Dai / Gregory D Sempowski / Kenneth Cronin / Robert Parks / Adam Yongxin Ye / Katayoun Mansouri / Maggie Barr / Novalia Pishesha / Aimee Chapdelaine Williams / Lucas Vieira Francisco / Anand Saminathan / Hanqin Peng / Himanshu Batra / Lorenza Bellusci / Surender Khurana / S Munir Alam / David C Montefiori / Kevin O Saunders / Ming Tian / Hidde Ploegh / Tom Kirchhausen / Bing Chen / Barton F Haynes / Frederick W Alt /  Abstract: SARS-CoV-2 Omicron subvariants have generated a worldwide health crisis due to resistance to most approved SARS-CoV-2 neutralizing antibodies and evasion of vaccination-induced antibodies. To manage ...SARS-CoV-2 Omicron subvariants have generated a worldwide health crisis due to resistance to most approved SARS-CoV-2 neutralizing antibodies and evasion of vaccination-induced antibodies. To manage Omicron subvariants and prepare for new ones, additional means of isolating broad and potent humanized SARS-CoV-2 neutralizing antibodies are desirable. Here, we describe a mouse model in which the primary B cell receptor (BCR) repertoire is generated solely through V(D)J recombination of a human V1-2 heavy chain (HC) and, substantially, a human Vκ1-33 light chain (LC). Thus, primary humanized BCR repertoire diversity in these mice derives from immensely diverse HC and LC antigen-contact CDR3 sequences generated by nontemplated junctional modifications during V(D)J recombination. Immunizing this mouse model with SARS-CoV-2 (Wuhan-Hu-1) spike protein immunogens elicited several V1-2/Vκ1-33-based neutralizing antibodies that bound RBD in a different mode from each other and from those of many prior patient-derived V1-2-based neutralizing antibodies. Of these, SP1-77 potently and broadly neutralized all SARS-CoV-2 variants through BA.5. Cryo-EM studies revealed that SP1-77 bound RBD away from the receptor-binding motif via a CDR3-dominated recognition mode. Lattice light-sheet microscopy-based studies showed that SP1-77 did not block ACE2-mediated viral attachment or endocytosis but rather blocked viral-host membrane fusion. The broad and potent SP1-77 neutralization activity and nontraditional mechanism of action suggest that it might have therapeutic potential. Likewise, the SP1-77 binding epitope may inform vaccine strategies. Last, the type of humanized mouse models that we have described may contribute to identifying therapeutic antibodies against future SARS-CoV-2 variants and other pathogens. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27046.map.gz emd_27046.map.gz | 14.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27046-v30.xml emd-27046-v30.xml emd-27046.xml emd-27046.xml | 28.8 KB 28.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27046_fsc.xml emd_27046_fsc.xml | 5.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_27046.png emd_27046.png | 67.1 KB | ||
| Others |  emd_27046_half_map_1.map.gz emd_27046_half_map_1.map.gz emd_27046_half_map_2.map.gz emd_27046_half_map_2.map.gz | 11.9 MB 11.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27046 http://ftp.pdbj.org/pub/emdb/structures/EMD-27046 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27046 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27046 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27046.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27046.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final sharpened map of negative stain electron microscopy single particle reconstruction of monoclonal antibody VHH7-5-82 in complex with SARS-CoV2 2P spike. | ||||||||||||||||||||
| Voxel size | X=Y=Z: 2.4 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half-map 1 of negative stain electron microscopy single...
| File | emd_27046_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 of negative stain electron microscopy single particle reconstruction of monoclonal antibody VHH7-5-82 in complex with SARS-CoV2 2P spike. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 2 of negative stain electron microscopy single...
| File | emd_27046_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 of negative stain electron microscopy single particle reconstruction of monoclonal antibody VHH7-5-82 in complex with SARS-CoV2 2P spike. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of SARS-CoV2 2P spike with VHH7-5-82 antibody Fab
| Entire | Name: Complex of SARS-CoV2 2P spike with VHH7-5-82 antibody Fab |
|---|---|
| Components |
|
-Supramolecule #1: Complex of SARS-CoV2 2P spike with VHH7-5-82 antibody Fab
| Supramolecule | Name: Complex of SARS-CoV2 2P spike with VHH7-5-82 antibody Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 48 KDa |
-Supramolecule #2: SARS-CoV2 2P spike
| Supramolecule | Name: SARS-CoV2 2P spike / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Severe acute respiratory syndrome coronavirus Severe acute respiratory syndrome coronavirus |
-Supramolecule #3: VHH7-5-82 Antibody Fab Light Chain, VHH7-5-82 Antibody Fab Heavy Chain
| Supramolecule | Name: VHH7-5-82 Antibody Fab Light Chain, VHH7-5-82 Antibody Fab Heavy Chain type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
-Macromolecule #1: SARS-CoV2 2P spike
| Macromolecule | Name: SARS-CoV2 2P spike / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Severe acute respiratory syndrome coronavirus Severe acute respiratory syndrome coronavirus |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRF DNPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP F LGVYYHKN NKSWMESEFR VYSSANNCTF ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRF DNPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP F LGVYYHKN NKSWMESEFR VYSSANNCTF EYVSQPFLMD LEGKQGNFKN LREFVFKNID GYFKIYSKHT PI NLVRDLP QGFSALEPLV DLPIGINITR FQTLLALHRS YLTPGDSSSG WTAGAAAYYV GYLQPRTFLL KYN ENGTIT DAVDCALDPL SETKCTLKSF TVEKGIYQTS NFRVQPTESI VRFPNITNLC PFGEVFNATR FASV YAWNR KRISNCVADY SVLYNSASFS TFKCYGVSPT KLNDLCFTNV YADSFVIRGD EVRQIAPGQT GKIAD YNYK LPDDFTGCVI AWNSNNLDSK VGGNYNYLYR LFRKSNLKPF ERDISTEIYQ AGSTPCNGVE GFNCYF PLQ SYGFQPTNGV GYQPYRVVVL SFELLHAPAT VCGPKKSTNL VKNKCVNFNF NGLTGTGVLT ESNKKFL PF QQFGRDIADT TDAVRDPQTL EILDITPCSF GGVSVITPGT NTSNQVAVLY QDVNCTEVPV AIHADQLT P TWRVYSTGSN VFQTRAGCLI GAEHVNNSYE CDIPIGAGIC ASYQTQTNSP GSASSVASQS IIAYTMSLG AENSVAYSNN SIAIPTNFTI SVTTEILPVS MTKTSVDCTM YICGDSTECS NLLLQYGSFC TQLNRALTGI AVEQDKNTQ EVFAQVKQIY KTPPIKDFGG FNFSQILPDP SKPSKRSFIE DLLFNKVTLA DAGFIKQYGD C LGDIAARD LICAQKFNGL TVLPPLLTDE MIAQYTSALL AGTITSGWTF GAGAALQIPF AMQMAYRFNG IG VTQNVLY ENQKLIANQF NSAIGKIQDS LSSTASALGK LQDVVNQNAQ ALNTLVKQLS SNFGAISSVL NDI LSRLDP PEAEVQIDRL ITGRLQSLQT YVTQQLIRAA EIRASANLAA TKMSECVLGQ SKRVDFCGKG YHLM SFPQS APHGVVFLHV TYVPAQEKNF TTAPAICHDG KAHFPREGVF VSNGTHWFVT QRNFYEPQII TTDNT FVSG NCDVVIGIVN NTVYDPLQPE LDSFKEELDK YFKNHTSPDV DLGDISGINA SVVNIQKEID RLNEVA KNL NESLIDLQEL GKYEQGSGYI PEAPRDGQAY VRKDGEWVLL STFLGRSLEV LFQGPGHHHH HHHHSAW SH PQFEKGGGSG GGGSGGSAWS HPQFEK |
-Macromolecule #2: VHH7-5-82 Antibody Fab Heavy Chain
| Macromolecule | Name: VHH7-5-82 Antibody Fab Heavy Chain / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGAE VKKPGASVKV SCKASGYTFT GYYIHWVRQA PGQGLEWMGW INPNSGGSHY AQKFQGRVTL TRDTSISTAY MELTRLRSDD TAVYYCARDR VGQRFPYWGQ GTLVTVSSAS TKGPSVFPLA PSSKSTSGGT AALGCLVKDY FPEPVTVSWN SGALTSGVHT ...String: QVQLVQSGAE VKKPGASVKV SCKASGYTFT GYYIHWVRQA PGQGLEWMGW INPNSGGSHY AQKFQGRVTL TRDTSISTAY MELTRLRSDD TAVYYCARDR VGQRFPYWGQ GTLVTVSSAS TKGPSVFPLA PSSKSTSGGT AALGCLVKDY FPEPVTVSWN SGALTSGVHT FPAVLQSSGL YSLSSVVTVP SSSLGTQTYI CNVNHKPSNT KVDKKVEPKS CDKTHT |
-Macromolecule #3: VHH7-5-82 Antibody Fab Light Chain
| Macromolecule | Name: VHH7-5-82 Antibody Fab Light Chain / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS LSASVGDRVT ITCQASQDIF NCLNWYQQKP GKAPKLLIYD ASNLETGVPS RFSGSGSGTD FTFTISSLQP EDVATYFCQQ YDNLWTFGGG TKLEIKRTVA APSVFIFPPS DEQLKSGTAS VVCLLNNFYP REAKVQWKVD NALQSGNSQE SVTEQDSKDS ...String: DIQMTQSPSS LSASVGDRVT ITCQASQDIF NCLNWYQQKP GKAPKLLIYD ASNLETGVPS RFSGSGSGTD FTFTISSLQP EDVATYFCQQ YDNLWTFGGG TKLEIKRTVA APSVFIFPPS DEQLKSGTAS VVCLLNNFYP REAKVQWKVD NALQSGNSQE SVTEQDSKDS TYSLSSTLTL SKADYEKHKV YACEVTHQGL SSPVTKSFNR GEC |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl formate Details: For negative staining, 5 microliter of the diluted and quenched sample was applied to a freshly glow-discharged, 300 mesh, carbon-coated EM grid and incubated 10-12 s, then rinsed with 70 ...Details: For negative staining, 5 microliter of the diluted and quenched sample was applied to a freshly glow-discharged, 300 mesh, carbon-coated EM grid and incubated 10-12 s, then rinsed with 70 microliter of 2% uranyl formate delivered in one smooth ejection from a pipet. The drop of uranyl formate remaining on the grid was allowed to incubate for 60 s, then the grid was blotted with filter paper and allowed to air dry. | ||||||||||||
| Grid | Model: Homemade / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 5 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||||||
| Details | To form the Fab-spike complex, spike was mixed with Fab at a 9:1 molar ratio of fab to spike and incubated at 37 degrees C for 1 h. Then complex was cross-linked by diluting to 200 micrograms/ml spike with 5 g/dl Glycerol in HEPES buffered saline (HBS), containing 20 mM HEPES pH 7.4 and 150 mM NaCl, augmented with 8 mM glutaraldehyde, then incubated 5 min and quenched by addition of sufficient 1 M Tris stock to give 75 mM final Tris concentration and incubated for an additional 5 min. |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS EM420 |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.2 µm / Nominal magnification: 49000 Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.2 µm / Nominal magnification: 49000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: OTHER / Digitization - Dimensions - Width: 8736 pixel / Digitization - Dimensions - Height: 8740 pixel / Number grids imaged: 1 / Number real images: 108 / Average exposure time: 0.25 sec. / Average electron dose: 32.0 e/Å2 Details: Images were collected with a minimal dose approach with the beam blanked except during initial grid orientation and image collection. To create an initial atlas, the entire grid was imaged ...Details: Images were collected with a minimal dose approach with the beam blanked except during initial grid orientation and image collection. To create an initial atlas, the entire grid was imaged once at 49x magnification at very low beam intensity by decreasing the emission current, strongly focusing C1 (Spot Size 6) and defocusing C2 (Intensity knob fully clockwise). The stage was then quickly translated to center the desired gridsquare, with an estimated exposure of < 2 s at this very low illumination level, at which point the beam was blanked. A 3,300x magnification was then selected so that a single gridsquare approximately fills the scintillator screen. The beam was then unblanked and the stage manually tilted and the eucentric height quickly adjusted by noting the movement of the gridbars, and then the stage was translated to center on the lower-left corner of the gridsquare, with an estimated exposure of < 2 s at this very low illumination level, at which point the beam was blanked. A magnfication of 49,000x was then selected and C2 focused by a known number of clicks of the Intensity knob so that the beam would approximately fill the scintillator screen. The beam was then unblanked, the objective lens focused by eye and then adjusted to approximately 0.5 micron underfocused, and C2 adjusted so the beam just filled the scintillator screen. Because this focusing step involved a long exposure to the focused beam, no data was collected in this area. For data collection, the stage was translated blindly, with the beam blanked, by noting the micrometer markings on the left-hand stage control, to move to an un-irradiated area that was approximately two screen-widths vertically away from the initial position. The beam was then unblanked, an image captured with 0.25 s exposure to the focused beam without any adjustment of the objective lens focus, and the beam then re-blanked. The FFT of the collected image was examined and the position of the first Thon ring noted and the objective lens focus adjusted if needed for subsequent images. The stage was then moved blindly in the same direction to an un-irradiated area and a new image collected. In this way images were collected until the gridbar was encountered, at which point the beam was unblanked and the stage moved so that the gridbar occluded all of the beam except a small sliver of the gridsquare at the bottom or top of the screen, and the right hand stage control was used to translate the specimen to a new location at least two screen widths away horizontally. In this way the entire gridsquare could be imaged in a rastered fashion with each area only irradiated with the full strength beam during the 0.25 s image capture. |
 Movie
Movie Controller
Controller









 Z
Z Y
Y X
X


















