-Search query
-Search result
Showing all 39 items for (author: rames & m)

EMDB-41062: 
CryoEM structure of an inward-facing MelBSt at a Na(+)-bound and sugar low-affinity conformation
Method: single particle / : Guan L

PDB-8t60: 
CryoEM structure of an inward-facing MelBSt at a Na(+)-bound and sugar low-affinity conformation
Method: single particle / : Guan L
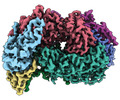
EMDB-29978: 
McrD binds asymmetrically to methyl-coenzyme M reductase improving active site accessibility during assembly
Method: single particle / : Joiner AMN, Chadwick GL, Nayak DD
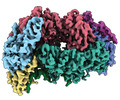
EMDB-29979: 
Apo-apo MCR assembly intermediate
Method: single particle / : Joiner AMN, Chadwick GL, Nayak DD

PDB-8gf5: 
McrD binds asymmetrically to methyl-coenzyme M reductase improving active site accessibility during assembly
Method: single particle / : Joiner AMN, Chadwick GL, Nayak DD

PDB-8gf6: 
Apo-apo MCR assembly intermediate
Method: single particle / : Joiner AMN, Chadwick GL, Nayak DD

EMDB-15073: 
Rabies virus glycoprotein in complex with Fab fragments of 17C7 and 1112-1 neutralizing antibodies
Method: single particle / : Ng WM, Fedosyuk S, English S, Augusto G, Berg A, Thorley L, Haselon AS, Segireddy RR, Bowden TA, Douglas AD

PDB-8a1e: 
Rabies virus glycoprotein in complex with Fab fragments of 17C7 and 1112-1 neutralizing antibodies
Method: single particle / : Ng WM, Fedosyuk S, English S, Augusto G, Berg A, Thorley L, Haselon AS, Segireddy RR, Bowden TA, Douglas AD

EMDB-33300: 
Cryo-EM structure of PEIP-Bs_enolase complex
Method: single particle / : Li S, Zhang K

EMDB-25399: 
Molecular mechanism of the the wake-promoting agent TAK-925
Method: single particle / : Yin J, Chapman K, Lian P, De Brabander JK, Rosenbaum DM

PDB-7sr8: 
Molecular mechanism of the the wake-promoting agent TAK-925
Method: single particle / : Yin J, Chapman K, Lian P, De Brabander JK, Rosenbaum DM

EMDB-25389: 
Structure of the orexin-2 receptor(OX2R) bound to TAK-925, Gi and scFv16
Method: single particle / : McGrath AP, Kang Y, Flinspach M

PDB-7sqo: 
Structure of the orexin-2 receptor(OX2R) bound to TAK-925, Gi and scFv16
Method: single particle / : McGrath AP, Kang Y, Flinspach M

EMDB-9401: 
CH505 SOSIP.664 trimer in complex with DH522.2 Fab
Method: single particle / : Fera D, Harrison SC

EMDB-8361: 
Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit
Method: single particle / : Liu Z, Gutierrez-Vargas C, Wei J, Grassucci RA, Ramesh M, Espina N, Sun M, Tutuncuoglu B, Madison-Antenucci S, Woolford Jr JL, Tong L, Frank J

PDB-5t5h: 
Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit
Method: single particle / : Liu Z, Gutierrez-Vargas C, Wei J, Grassucci RA, Ramesh M, Espina N, Sun M, Tutuncuoglu B, Madison-Antenucci S, Woolford Jr JL, Tong L, Frank J

EMDB-6041: 
Imaging proteins and cells in liquid solution at nanometer resolution by electron microscopy
Method: electron tomography / : Ren G, Peng B, Zhang L, Lei D, Lu Z, Wong E, Zhang M, Rames MJ, Liu J, De Yoreo JJ, Zhang J

EMDB-6042: 
Imaging proteins and cells in liquid solution at nanometer resolution by electron microscopy
Method: electron tomography / : Ren G, Peng B, Zhang L, Lei D, Lu Z, Wong E, Zhang M, Rames MJ, Liu J, De Yoreo JJ, Zhang J

EMDB-6099: 
Exploring IgG1 Antibody Conformational Flexibility in Three Dimensions
Method: electron tomography / : Zhang X, Zhang L, Tong H, Peng B, Rames MJ, Zhang S, Ren G

EMDB-5933: 
Cryo-EM structure of Dengue serotype 3 virus at 28 degrees C
Method: single particle / : Fibriansah G, Tan JL, Smith SA, de Alwis R, Ng TS, Kostyuchenko VA, Kukkaro P, de Silva AM, Crowe Jr JE, Lok SM

EMDB-5934: 
Cryo-EM structure of Dengue serotype 3 virus at 37 degrees C
Method: single particle / : Fibriansah G, Tan JL, Smith SA, de Alwis R, Ng TS, Kostyuchenko VA, Kukkaro P, de Silva AM, Crowe Jr JE, Lok SM

EMDB-5935: 
Cryo-EM structure of Dengue virus serotype 3 in complex with human antibody 5J7 Fab
Method: single particle / : Fibriansah G, Tan JL, Smith SA, de Alwis R, Ng TS, Kostyuchenko VA, Kukkaro P, de Silva AM, Crowe Jr JE, Lok SM

PDB-3j6s: 
Cryo-EM structure of Dengue virus serotype 3 at 28 degrees C
Method: single particle / : Fibriansah G, Tan JL, Smith SA, de Alwis R, Ng TS, Kostyuchenko VA, Kukkaro P, de Silva AM, Crowe Jr JE, Lok SM

PDB-3j6t: 
Cryo-EM structure of Dengue virus serotype 3 at 37 degrees C
Method: single particle / : Fibriansah G, Tan JL, Smith SA, de Alwis R, Ng TS, Kostyuchenko VA, Kukkaro P, de Silva AM, Crowe Jr JE, Lok SM

PDB-3j6u: 
Cryo-EM structure of Dengue virus serotype 3 in complex with human antibody 5J7 Fab
Method: single particle / : Fibriansah G, Tan JL, Smith SA, de Alwis R, Ng TS, Kostyuchenko VA, Kukkaro P, de Silva AM, Crowe Jr JE, Lok SM

EMDB-5759: 
Hepatitis C Virus E2 Envelope Glycoprotein Core Structure
Method: single particle / : Nieusma T, Kong L, Giang E, Kadam RU, Cogburn KE, Hua Y, Dai X, Stanfield RL, Burton DR, Wilson IA, Law M, Ward AB

EMDB-5760: 
Hepatitis C Virus E2 Envelope Glycoprotein Core Structure
Method: single particle / : Nieusma T, Kong L, Giang E, Kadam RU, Cogburn KE, Hua Y, Dai X, Stanfield RL, Burton DR, Wilson IA, Law M, Ward AB

EMDB-5761: 
Hepatitis C Virus E2 Envelope Glycoprotein Core Structure
Method: single particle / : Nieusma T, Kong L, Giang E, Kadam RU, Cogburn KE, Hua Y, Dai X, Stanfield RL, Burton DR, Wilson IA, Law M, Ward AB
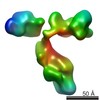
EMDB-2285: 
3D map of a peptide conjugated antibody particle by individual-particle electron tomography (IPET) and optimized negative-staining.
Method: electron tomography / : Tong H, Zhang L, Kaspar A, Rames M, Huang L, Woodnutt G, Ren G

EMDB-2286: 
3D map of another peptide conjugated antibody particle by individual-particle electron tomography (IPET) and optimized negative-staining.
Method: electron tomography / : Tong H, Zhang L, Kaspar A, Rames M, Huang L, Woodnutt G, Ren G

EMDB-2138: 
EM map of influenza H1 hemagglutatin in complex with C05 Fab
Method: single particle / : Khayat R, Lee JH, Ekiert DC, Wilson IA, Ward AB

EMDB-2139: 
EM map of an influenza H3 hemagglutatin in complex with C05 Fab
Method: single particle / : Khayat R, Lee JH, Ekiert DC, Wilson IA, Ward AB

EMDB-2140: 
EM map of influenza H2 hemagglutatin in complex with C05 Fab
Method: single particle / : Khayat R, Lee JH, Ekiert DC, Wilson IA, Ward AB

EMDB-1612: 
Nanoscale DNA box
Method: single particle / : Andersen ES, Dong M, Nielsen MM, Jahn K, Subramani R, Mamdouh W, Golas MM, Sander B, Stark H, Oliveira CL, Pedersen JS, Birkedal V, Besenbacher F, Gothelf KV, Kjems J

EMDB-1109: 
Structural insights into the activity of enhancer-binding proteins.
Method: single particle / : Rappas M, Schumacher J, Beuron F, Niwa H, Bordes P, Wigneshweraraj S, Keetch CA, Robinson CV, Buck M, Zhang X
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
