-Search query
-Search result
Showing 1 - 50 of 1,092 items for (author: o & ryan & l)

EMDB-43931: 
CryoEM structure of activated CRAF/MEK/14-3-3 complex with NST-628
Method: single particle / : Quade B, Cohen SE, Huang X

EMDB-43932: 
Activated CRAF/MEK heterotetramer from focused refinement of CRAF/MEK/14-3-3 complex
Method: single particle / : Quade B, Cohen SE, Huang X

PDB-9axa: 
CryoEM structure of activated CRAF/MEK/14-3-3 complex with NST-628
Method: single particle / : Quade B, Cohen SE, Huang X

PDB-9axc: 
Activated CRAF/MEK heterotetramer from focused refinement of CRAF/MEK/14-3-3 complex
Method: single particle / : Quade B, Cohen SE, Huang X

EMDB-42392: 
Cryo-EM Structure of the Helicobacter pylori cagYdAP PR
Method: single particle / : Roberts JR

EMDB-42290: 
Cryo-EM Structure of the Helicobacter pylori CagYdAP OMC
Method: single particle / : Roberts JR

EMDB-42393: 
Cryo-EM Structure of the Helicobacter pylori dcagM PR
Method: single particle / : Roberts JR

EMDB-42395: 
Cryo-EM Structure of the Helicobacter pylori dcagT PR
Method: single particle / : Roberts JR
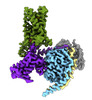
EMDB-43647: 
CryoEM structure of Gi-coupled TAS2R14 with cholesterol and an intracellular tastant
Method: single particle / : Kim Y, Gumpper RH, Roth BL
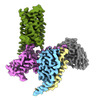
EMDB-43650: 
CryoEM structure of Ggust-coupled TAS2R14 with cholesterol and an intracellular tastant
Method: single particle / : Kim Y, Gumpper RH, Roth BL

EMDB-43656: 
CryoEM structure of Gi-coupled TAS2R14 with cholesterol and an intracellular tastant (Locally refined map)
Method: single particle / : Kim Y, Gumpper RH, Roth BL

EMDB-43657: 
CryoEM structure of Ggust-coupled TAS2R14 with cholesterol and an intracellular tastant (Locally refined map)
Method: single particle / : Kim Y, Gumpper RH, Roth BL

PDB-8vy7: 
CryoEM structure of Gi-coupled TAS2R14 with cholesterol and an intracellular tastant
Method: single particle / : Kim Y, Gumpper RH, Roth BL

PDB-8vy9: 
CryoEM structure of Ggust-coupled TAS2R14 with cholesterol and an intracellular tastant
Method: single particle / : Kim Y, Gumpper RH, Roth BL

EMDB-41907: 
Computationally Designed, Expandable O4 Octahedral Handshake Nanocage
Method: single particle / : Weidle C, Borst A
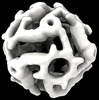
EMDB-42031: 
Computational Designed Nanocage O43_129_+8
Method: single particle / : Weidle C, Kibler RD

EMDB-43318: 
Twistless helix 12 repeat ring design R12B
Method: single particle / : Calise SJ, Kollman JM

EMDB-29974: 
Cryo-EM structure of synthetic tetrameric building block sC4
Method: single particle / : Redler RL, Huddy TF, Hsia Y, Baker D, Ekiert D, Bhabha G

EMDB-41364: 
CryoEM Structure of a Computationally Designed T3 Tetrahedral Nanocage
Method: single particle / : Weidle C, Borst AJ

EMDB-42906: 
Computational Designed Nanocage O43_129
Method: single particle / : Weidle C, Kibler RD

EMDB-42944: 
Computational Designed Nanocage O43_129_+4
Method: single particle / : Carr KD, Weidle C, Borst AJ

PDB-8gel: 
Cryo-EM structure of synthetic tetrameric building block sC4
Method: single particle / : Redler RL, Huddy TF, Hsia Y, Baker D, Ekiert D, Bhabha G

PDB-8tl7: 
CryoEM Structure of a Computationally Designed T3 Tetrahedral Nanocage
Method: single particle / : Weidle C, Borst AJ

PDB-8v3b: 
Computational Designed Nanocage O43_129_+4
Method: single particle / : Carr KD, Weidle C, Borst AJ

EMDB-42681: 
The structure of the native cardiac thin filament troponin core in Ca2+-free state from the upper strand
Method: single particle / : Galkin VE, Risi CM

EMDB-42682: 
The structure of the native cardiac thin filament troponin core in Ca2+-free tilted state from the upper strand
Method: single particle / : Galkin VE, Risi CM

EMDB-42683: 
The structure of the native cardiac thin filament troponin core in Ca2+-free rotated state from the upper strand
Method: single particle / : Galkin VE, Risi CM

EMDB-42800: 
The structure of the native cardiac thin filament troponin core in Ca2+-free state from the lower strand
Method: single particle / : Galkin VE, Risi CM

EMDB-42833: 
The structure of the native cardiac thin filament troponin core in Ca2+-free rotated state from the lower strand
Method: single particle / : Galkin VE, Risi CM

EMDB-42835: 
The structure of the native cardiac thin filament troponin core in Ca2+-free tilted state from the lower strand
Method: single particle / : Galkin VE, Risi CM

EMDB-42846: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound fully activated state from the upper strand
Method: single particle / : Galkin VE, Risi CM

EMDB-42847: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound partially activated state from the upper strand
Method: single particle / : Galkin VE, Risi CM

EMDB-42849: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound fully activated state 1 from the lower strand
Method: single particle / : Galkin VE, Risi CM

EMDB-42856: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound fully activated state 2 from the lower strand
Method: single particle / : Galkin VE, Risi CM

EMDB-42858: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound partially activated state from the lower strand
Method: single particle / : Galkin VE, Risi CM

EMDB-42874: 
The structure of the native cardiac thin filament troponin core in Ca2+-free state from the upper strand activated by the C1-domain of cardiac myosin binding protein C
Method: single particle / : Galkin VE, Risi CM

PDB-8uww: 
The structure of the native cardiac thin filament troponin core in Ca2+-free state from the upper strand
Method: single particle / : Galkin VE, Risi CM

PDB-8uwx: 
The structure of the native cardiac thin filament troponin core in Ca2+-free tilted state from the upper strand
Method: single particle / : Galkin VE, Risi CM

PDB-8uwy: 
The structure of the native cardiac thin filament troponin core in Ca2+-free rotated state from the upper strand
Method: single particle / : Galkin VE, Risi CM

PDB-8uyd: 
The structure of the native cardiac thin filament troponin core in Ca2+-free state from the lower strand
Method: single particle / : Galkin VE, Risi CM

PDB-8uz5: 
The structure of the native cardiac thin filament troponin core in Ca2+-free rotated state from the lower strand
Method: single particle / : Galkin VE, Risi CM

PDB-8uz6: 
The structure of the native cardiac thin filament troponin core in Ca2+-free tilted state from the lower strand
Method: single particle / : Galkin VE, Risi CM

PDB-8uzx: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound fully activated state from the upper strand
Method: single particle / : Galkin VE, Risi CM

PDB-8uzy: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound partially activated state from the upper strand
Method: single particle / : Galkin VE, Risi CM

PDB-8v01: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound fully activated state 1 from the lower strand
Method: single particle / : Galkin VE, Risi CM

PDB-8v0i: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound fully activated state 2 from the lower strand
Method: single particle / : Galkin VE, Risi CM

PDB-8v0k: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound partially activated state from the lower strand
Method: single particle / : Galkin VE, Risi CM

PDB-8v0y: 
The structure of the native cardiac thin filament troponin core in Ca2+-free state from the upper strand activated by the C1-domain of cardiac myosin binding protein C
Method: single particle / : Galkin VE, Risi CM

EMDB-42526: 
Alpha7-nicotinic acetylcholine receptor bound to epibatidine
Method: single particle / : Burke SM, Hibbs RE, Noviello CM
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
