-Search query
-Search result
Showing 1 - 50 of 417 items for (author: low & j)
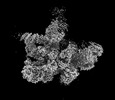
EMDB-18920: 
Plastid-encoded RNA polymerase (consensus map)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A
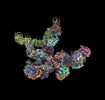
EMDB-18935: 
Plastid-encoded RNA polymerase
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18952: 
Plastid-encoded RNA polymerase transcription elongation complex (consensus map)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18964: 
Plastid-encoded RNA polymerase (Region 1)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18965: 
Plastid-encoded RNA polymerase (Region 2)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18974: 
Plastid-encoded RNA polymerase (Region 3)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18975: 
Plastid-encoded RNA polymerase (Region 4)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18976: 
Plastid-encoded RNA polymerase (Region 5)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18982: 
Plastid-encoded RNA polymerase (Region 6)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18983: 
Plastid-encoded RNA polymerase (Region 8)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18985: 
Plastid-encoded RNA polymerase (Region 9)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18986: 
Plastid-encoded RNA polymerase transcription elongation complex (Region 1)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18995: 
Plastid-encoded RNA polymerase transcription elongation complex (Region 2)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18996: 
Plastid-encoded RNA polymerase transcription elongation complex (Region 3)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18998: 
Plastid-encoded RNA polymerase (Region 7)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-19007: 
Plastid-encoded RNA polymerase transcription elongation complex (Region 4)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A
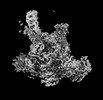
EMDB-19010: 
Plastid-encoded RNA polymerase transcription elongation complex (PAP2-mRNA)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

PDB-8r5o: 
Plastid-encoded RNA polymerase
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

PDB-8r6s: 
Plastid-encoded RNA polymerase (Integrated model)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

PDB-8rdj: 
Plastid-encoded RNA polymerase transcription elongation complex (Integrated model)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A
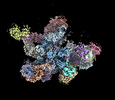
EMDB-19023: 
Plastid-encoded RNA polymerase transcription elongation complex
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

PDB-8ras: 
Plastid-encoded RNA polymerase transcription elongation complex
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-36480: 
CryoEM structure of isNS1 in complex with Fab56.2 and HDL
Method: single particle / : Chew BLA, Luo D

EMDB-36483: 
CryoEM structure of isNS1 in complex with Fab56.2
Method: single particle / : Chew BLA, Luo D
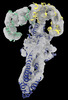
EMDB-18149: 
GBP1 bound by 14-3-3sigma
Method: single particle / : Pfleiderer MM, Liu X, Fisch D, Anastasakou E, Frickel EM, Galej WP

PDB-8q4l: 
GBP1 bound by 14-3-3sigma
Method: single particle / : Pfleiderer MM, Liu X, Fisch D, Anastasakou E, Frickel EM, Galej WP

EMDB-16758: 
PHT1 in the outward facing conformation, bound to Sb27
Method: single particle / : Custodio T, Killer M, Loew C

EMDB-41153: 
Integrin alpha-v beta-8 in complex with minibinder B8_BP_dsulf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

EMDB-41154: 
Integrin alpha-v beta-6 in complex with minibinder B6_BP_dslf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

PDB-8tcf: 
Integrin alpha-v beta-8 in complex with minibinder B8_BP_dsulf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

PDB-8tcg: 
Integrin alpha-v beta-6 in complex with minibinder B6_BP_dslf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

EMDB-15395: 
Cryo-EM structure of crescentin filaments (stutter mutant, C1 symmetry and small box)
Method: single particle / : Liu Y, Lowe J

EMDB-15398: 
Cryo-EM structure of crescentin filaments (stutter mutant, C2, symmetry and small box)
Method: single particle / : Liu Y, Lowe J

EMDB-15399: 
Cryo-EM map of crescentin filament in complex with a megabody (stutter mutant, C2 symmetry, large box)
Method: single particle / : Liu Y, Lowe J

EMDB-15400: 
Cryo-EM map of crescentin filaments in complex with a megabody (wildtype, C2, large box)
Method: single particle / : Liu Y, Lowe J

EMDB-15401: 
Cryo-EM structure of crescentin filaments (wildtype, C1 symmetry and small box)
Method: single particle / : Liu Y, Lowe J

EMDB-15402: 
Cryo-EM structure of crescentin filaments (wildtype, C2 symmetry and small box)
Method: single particle / : Liu Y, Lowe J

EMDB-15446: 
Cryo-EM structure of crescentin filaments (stutter mutant, C1 symmetry and large box)
Method: single particle / : Liu Y, Lowe J

EMDB-15465: 
Cryo-EM structure of crescentin filaments (wildtype, C1 symmetry and large box)
Method: single particle / : Liu Y, Lowe J

EMDB-15473: 
Cryo-EM structure of crescentin filaments (wildtype, C2 symmetry and large box)
Method: single particle / : Liu Y, Lowe J

EMDB-15476: 
Cryo-EM structure of crescentin filaments (stutter mutant, C2 symmetry and large box)
Method: single particle / : Liu Y, Lowe J

PDB-8afe: 
Cryo-EM structure of crescentin filaments (stutter mutant, C1 symmetry and small box)
Method: single particle / : Liu Y, Lowe J

PDB-8afh: 
Cryo-EM structure of crescentin filaments (stutter mutant, C2, symmetry and small box)
Method: single particle / : Liu Y, Lowe J

PDB-8afl: 
Cryo-EM structure of crescentin filaments (wildtype, C1 symmetry and small box)
Method: single particle / : Liu Y, Lowe J

PDB-8afm: 
Cryo-EM structure of crescentin filaments (wildtype, C2 symmetry and small box)
Method: single particle / : Liu Y, Lowe J

PDB-8ahl: 
Cryo-EM structure of crescentin filaments (stutter mutant, C1 symmetry and large box)
Method: single particle / : Liu Y, Lowe J

PDB-8aia: 
Cryo-EM structure of crescentin filaments (wildtype, C1 symmetry and large box)
Method: single particle / : Liu Y, Lowe J

PDB-8aix: 
Cryo-EM structure of crescentin filaments (wildtype, C2 symmetry and large box)
Method: single particle / : Liu Y, Lowe J

PDB-8ajb: 
Cryo-EM structure of crescentin filaments (stutter mutant, C2 symmetry and large box)
Method: single particle / : Liu Y, Lowe J

EMDB-33942: 
Cryo-EM structure of MERS-CoV spike protein, Two RBD-up conformation 2
Method: single particle / : Hsu STD, Chang NE, Weng ZW, Yang TJ, Draczkowski P
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
