-Search query
-Search result
Showing 1 - 50 of 878 items for (author: jian & ll)

EMDB-40411: 
PHF Tau from Down Syndrome
Method: helical / : Hoq MR, Bharath SR, Jiang W, Vago FS
EMDB-40413: 
SF Tau from Down Syndrome
Method: helical / : Hoq MR, Bharath SR, Jiang W, Vago FS
EMDB-40416: 
Type I beta-amyloid 42 Filaments from Down syndrome
Method: helical / : Hoq MR, Bharath SR, Vago FS, Jiang W

EMDB-40419: 
Type IIIa beta-amyloid 40 Filaments from Down syndrome
Method: helical / : Hoq MR, Vago FS, Bharath SR, Jiang W
EMDB-40421: 
Type IIIb beta-amyloid 40 Filaments from Down Syndrome
Method: helical / : Hoq MR, Vago FS, Bharath SR, Jiang W
PDB-8seh: 
PHF Tau from Down Syndrome
Method: helical / : Hoq MR, Bharath SR, Jiang W, Vago FS
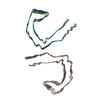
PDB-8sei: 
SF Tau from Down Syndrome
Method: helical / : Hoq MR, Bharath SR, Jiang W, Vago FS, Bharath SR

PDB-8sej: 
Type I beta-amyloid 42 Filaments from Down syndrome
Method: helical / : Hoq MR, Bharath SR, Vago FS, Jiang W

PDB-8sek: 
Type IIIa beta-amyloid 40 Filaments from Down syndrome
Method: helical / : Hoq MR, Vago FS, Bharath SR, Jiang W

PDB-8sel: 
Type IIIb beta-amyloid 40 Filaments from Down Syndrome
Method: helical / : Hoq MR, Vago FS, Bharath SR, Jiang W

EMDB-18267: 
Structure of the human 20S U5 snRNP core
Method: single particle / : Schneider S, Galej WP

EMDB-19041: 
Structure of the human 20S U5 snRNP
Method: single particle / : Schneider S, Galej WP

PDB-8q91: 
Structure of the human 20S U5 snRNP core
Method: single particle / : Schneider S, Galej WP

PDB-8rc0: 
Structure of the human 20S U5 snRNP
Method: single particle / : Schneider S, Galej WP

EMDB-42601: 
CryoEM structure of Kappa Opioid Receptor bound to a semi-peptide and Gi1
Method: single particle / : Fay JF, Che T
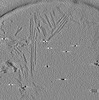
EMDB-28752: 
Cryo-electron tomography of wild type a-synuclein preformed fibrils
Method: electron tomography / : Jiang J, Boparai N, Dai W, Kim YS
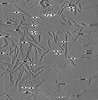
EMDB-28753: 
Cryo-electron tomography of S42Y a-synuclein preformed fibrils
Method: electron tomography / : Jiang J, Boparai N, Dai W, Kim YS

EMDB-29794: 
SARS-CoV-2 spike/Nb2 complex with 1 RBD up (local refinement at 5.6 A)
Method: single particle / : Ye G, Bu F, Liu B, Li F

EMDB-29795: 
SARS-CoV-2 spike/Nb3 complex with 2 RBDs up and 3 Nb3 bound at 2.5 A
Method: single particle / : Ye G, Bu F, Liu B, Li F

EMDB-29796: 
SARS-CoV-2 spike/Nb3 complex with 1 RBD up and 2 Nb3
Method: single particle / : Ye G, Bu F, Liu B, Li F

EMDB-29797: 
SARS-CoV-2 spike/Nb4 complex with 2 RBDs up and 3 Nb4 bound
Method: single particle / : Ye G, Bu F, Liu B, Li F

PDB-8g72: 
SARS-CoV-2 spike/Nb2 complex with 1 RBD up (local refinement at 5.6 A)
Method: single particle / : Ye G, Bu F, Liu B, Li F

PDB-8g73: 
SARS-CoV-2 spike/Nb3 complex with 2 RBDs up and 3 Nb3 bound at 2.5 A
Method: single particle / : Ye G, Bu F, Liu B, Li F

PDB-8g74: 
SARS-CoV-2 spike/Nb3 complex with 1 RBD up and 2 Nb3
Method: single particle / : Ye G, Bu F, Liu B, Li F

PDB-8g75: 
SARS-CoV-2 spike/Nb4 complex with 2 RBDs up and 3 Nb4 bound
Method: single particle / : Ye G, Bu F, Liu B, Li F

EMDB-36229: 
CryoEM structure of Gi-coupled MRGPRX1 with peptide agonist CNF-Tx2
Method: single particle / : Sun JP, Xu HE, Yang F, Liu ZM, Guo LL, Zhang YM, Fang GX, Tie L, Zhuang YM, Xue CY

EMDB-36232: 
CryoEM structure of Gq-coupled MRGPRX1 with peptide agonist BAM8-22
Method: single particle / : Sun JP, Xu HE, Yang F, Liu ZM, Guo LL, Zhang YM, Fang GX, Tie L, Zhuang YM, Xue CY

EMDB-36233: 
CryoEM structure of Gi-coupled MRGPRX1 with peptide agonist BAM8-22
Method: single particle / : Sun JP, Xu HE, Yang F, Liu ZM, Guo LL, Zhang YM, Fang GX, Tie L, Zhuang YM, Xue CY

EMDB-41048: 
Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5
Method: single particle / : Gorman J, Kwong PD

PDB-8t5c: 
Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5
Method: single particle / : Gorman J, Kwong PD

EMDB-37429: 
Cryo-EM structure of the SEP363856-bound mTAAR1-Gs complex
Method: single particle / : Rong NK, Guo LL, Zhang MH, Li Q, Yang F, Sun JP

EMDB-37430: 
Cryo-EM structure of the ZH8651-bound mTAAR1-Gs complex
Method: single particle / : Rong NK, Guo LL, Zhang MH, Li Q, Yang F, Sun JP

EMDB-37431: 
Cryo-EM structure of the TMA-bound mTAAR1-Gs complex
Method: single particle / : Rong NK, Guo LL, Zhang MH, Li Q, Yang F, Sun JP

EMDB-37432: 
Cryo-EM structure of the PEA-bound mTAAR1-Gs complex
Method: single particle / : Rong NK, Guo LL, Zhang MH, Li Q, Yang F, Sun JP

EMDB-37433: 
Cryo-EM structure of the ZH8667-bound mTAAR1-Gs complex
Method: single particle / : Rong NK, Guo LL, Zhang MH, Li Q, Yang F, Sun JP

EMDB-37434: 
Cryo-EM structure of the ZH8651-bound hTAAR1-Gs complex
Method: single particle / : Rong NK, Guo LL, Zhang MH, Li Q, Yang F, Sun JP

EMDB-37435: 
Cryo-EM structure of the ZH8651-bound mTAAR1-Gq complex
Method: single particle / : Rong NK, Guo LL, Zhang MH, Li Q, Yang F, Sun JP

EMDB-37436: 
Cryo-EM structure of the PEA-bound hTAAR1-Gs complex
Method: single particle / : Rong NK, Guo LL, Zhang MH, Li Q, Yang F, Sun JP

EMDB-37437: 
Cryo-EM structure of the CHA-bound mTAAR1-Gq complex
Method: single particle / : Rong NK, Guo LL, Zhang MH, Li Q, Yang F, Sun JP

EMDB-37438: 
Cryo-EM structure of the CHA-bound mTAAR1 complex
Method: single particle / : Rong NK, Guo LL, Zhang MH, Li Q, Yang F, Sun JP

EMDB-35987: 
Structural insights into photosystem II supercomplex and trimeric FCP antennae of a centric diatom Cyclotella meneghiniana
Method: single particle / : Shen LL, Li ZH, Shen JR, Wang WD

EMDB-36054: 
Structure of FCP trimer in Cyclotella meneghiniana
Method: single particle / : Shen LL, Li ZH, Shen JR, Wang WD

EMDB-37267: 
Structure of PSII-FCPII-G/H complex in the PSII-FCPII supercomplex from Cyclotella meneghiniana
Method: single particle / : Shen LL, Li ZH, Shen JR, Wang WD

EMDB-37268: 
Structure of PSII-FCPII-I/J/K complex in the PSII-FCPII supercomplex from Cyclotella meneghiniana
Method: single particle / : Shen LL, Li ZH, Shen JR, Wang WD

EMDB-42399: 
Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp and DNA
Method: single particle / : Huang Y, Marcus K, Subramanian S, Gee LC, Gorday K, Ghaffari-Kashani S, Luo X, Zhang L, O'Donnell M, Kuriyan J

EMDB-42402: 
Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp
Method: single particle / : Huang Y, Marcus K, Subramanian S, Gee LC, Gorday K, Ghaffari-Kashani S, Luo X, Zhang L, O'Donnell M, Kuriyan J

PDB-8unf: 
Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp and DNA
Method: single particle / : Huang Y, Marcus K, Subramanian S, Gee LC, Gorday K, Ghaffari-Kashani S, Luo X, Zhang L, O'Donnell M, Subramanian S, Kuriyan J

PDB-8unh: 
Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp
Method: single particle / : Huang Y, Marcus K, Subramanian S, Gee LC, Gorday K, Ghaffari-Kashani S, Luo X, Zhang L, O'Donnell M, Subramanian S, Kuriyan J

EMDB-37832: 
XBB.1.5.10 RBD in complex with ACE2
Method: single particle / : Feng LL, Feng L

EMDB-37835: 
XBB.1.5.70 spike protein in complex with ACE2
Method: single particle / : Feng LL
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
