[English] 日本語
 Yorodumi
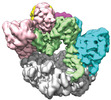
Yorodumi- EMDB-42402: Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | autoinhibited / DNA-free / catalytically inactive / stable /  REPLICATION REPLICATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationDNA clamp loader activity / bidirectional double-stranded viral DNA replication / viral DNA genome replication / DNA polymerase processivity factor activity / viral transcription /  Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement /  DNA replication / DNA replication /  ATP hydrolysis activity / ATP hydrolysis activity /  DNA binding / DNA binding /  ATP binding ATP bindingSimilarity search - Function | |||||||||
| Biological species |   Tequatrovirus T4 Tequatrovirus T4 | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.21 Å cryo EM / Resolution: 3.21 Å | |||||||||
 Authors Authors | Huang Y / Marcus K / Subramanian S / Gee LC / Gorday K / Ghaffari-Kashani S / Luo X / Zhang L / O'Donnell M / Kuriyan J | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM. Authors: Kendra Marcus / Yongjian Huang / Subu Subramanian / Christine L Gee / Kent Gorday / Sam Ghaffari-Kashani / Xiao Ran Luo / Lisa Zheng / Michael O'Donnell / Sriram Subramaniam / John Kuriyan /   Abstract: Clamp loaders are AAA+ ATPases that facilitate high-speed DNA replication. In eukaryotic and bacteriophage clamp loaders, ATP hydrolysis requires interactions between aspartate residues in one ...Clamp loaders are AAA+ ATPases that facilitate high-speed DNA replication. In eukaryotic and bacteriophage clamp loaders, ATP hydrolysis requires interactions between aspartate residues in one protomer, present in conserved 'DEAD-box' motifs, and arginine residues in adjacent protomers. We show that functional defects resulting from a DEAD-box mutation in the T4 bacteriophage clamp loader can be compensated by widely distributed single mutations in the ATPase domain. Using cryo-EM, we discovered an unsuspected inactive conformation of the clamp loader, in which DNA binding is blocked and the catalytic sites are disassembled. Mutations that restore function map to regions of conformational change upon activation, suggesting that these mutations may increase DNA affinity by altering the energetic balance between inactive and active states. Our results show that there are extensive opportunities for evolution to improve catalytic efficiency when an inactive intermediate is involved. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42402.map.gz emd_42402.map.gz | 15.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42402-v30.xml emd-42402-v30.xml emd-42402.xml emd-42402.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_42402.png emd_42402.png | 159.6 KB | ||
| Filedesc metadata |  emd-42402.cif.gz emd-42402.cif.gz | 6.5 KB | ||
| Others |  emd_42402_half_map_1.map.gz emd_42402_half_map_1.map.gz emd_42402_half_map_2.map.gz emd_42402_half_map_2.map.gz | 28.3 MB 28.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42402 http://ftp.pdbj.org/pub/emdb/structures/EMD-42402 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42402 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42402 | HTTPS FTP |
-Related structure data
| Related structure data |  8unhMC  8uh7C  8uk9C  8unfC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42402.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42402.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.048 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_42402_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
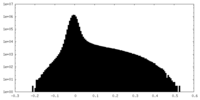
| Projections & Slices |
| ||||||||||||
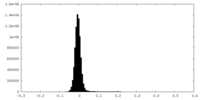
| Density Histograms |
-Half map: #1
| File | emd_42402_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
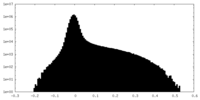
| Projections & Slices |
| ||||||||||||
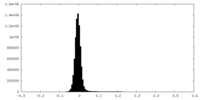
| Density Histograms |
- Sample components
Sample components
-Entire : T4 Bacteriophage Clamp Loader with Sliding Clamp
| Entire | Name: T4 Bacteriophage Clamp Loader with Sliding Clamp |
|---|---|
| Components |
|
-Supramolecule #1: T4 Bacteriophage Clamp Loader with Sliding Clamp
| Supramolecule | Name: T4 Bacteriophage Clamp Loader with Sliding Clamp / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Tequatrovirus T4 Tequatrovirus T4 |
-Supramolecule #2: T4 Bacteriophage Sliding Clamp (gp45)
| Supramolecule | Name: T4 Bacteriophage Sliding Clamp (gp45) / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:   Tequatrovirus T4 Tequatrovirus T4 |
-Supramolecule #3: T4 Bacteriophage Clamp Loader (gp44 + gp62)
| Supramolecule | Name: T4 Bacteriophage Clamp Loader (gp44 + gp62) / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Tequatrovirus T4 Tequatrovirus T4 |
-Macromolecule #1: Sliding-clamp-loader large subunit
| Macromolecule | Name: Sliding-clamp-loader large subunit / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Tequatrovirus T4 Tequatrovirus T4 |
| Molecular weight | Theoretical: 35.834254 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MITVNEKEHI LEQKYRPSTI DECILPAFDK ETFKSITSKG KIPHIILHSP SPGTGKTTVA KALCHDVNAD MMFVNGSDCK IDFVRGPLT NFASAASFDG RQKVIVIDEF DRSGLAESQR HLRSFMEAYS SNCSIIITAN NIDGIIKPLQ SRCRVITFGQ P TDEDKIEM ...String: MITVNEKEHI LEQKYRPSTI DECILPAFDK ETFKSITSKG KIPHIILHSP SPGTGKTTVA KALCHDVNAD MMFVNGSDCK IDFVRGPLT NFASAASFDG RQKVIVIDEF DRSGLAESQR HLRSFMEAYS SNCSIIITAN NIDGIIKPLQ SRCRVITFGQ P TDEDKIEM MKQMIRRLTE ICKHEGIAIA DMKVVAALVK KNFPDFRKTI GELDSYSSKG VLDAGILSLV TNDRGAIDDV LE SLKNKDV KQLRALAPKY AADYSWFVGK LAEEIYSRVT PQSIIRMYEI VGENNQYHGI AANTELHLAY LFIQLACEMQ WK UniProtKB: Sliding-clamp-loader large subunit |
-Macromolecule #2: Sliding-clamp-loader small subunit
| Macromolecule | Name: Sliding-clamp-loader small subunit / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Tequatrovirus T4 Tequatrovirus T4 |
| Molecular weight | Theoretical: 21.391703 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MSLFKDDIQL NEHQVAWYSK DWTAVQSAAD SFKEKAENEF FEIIGAINNK TKCSIAQKDY SKFMVENALS QFPECMPAVY AMNLIGSGL SDEAHFNYLM AAVPRGKRYG KWAKLVEDST EVLIIKLLAK RYQVNTNDAI NYKSILTKNG KLPLVLKELK G LVTDDFLK EVTKNVKEQK QLKKLALEW UniProtKB: Sliding-clamp-loader small subunit |
-Macromolecule #3: Sliding clamp
| Macromolecule | Name: Sliding clamp / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Tequatrovirus T4 Tequatrovirus T4 |
| Molecular weight | Theoretical: 24.881223 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MKLSKDTTAL LKNFATINSG IMLKSGQFIM TRAVNGTTYA EANISDVIDF DVAIYDLNGF LGILSLVNDD AEISQSEDGN IKIADARST IFWPAADPST VVAPNKPIPF PVASAVTEIK AEDLQQLLRV SRGLQIDTIA ITVKEGKIVI NGFNKVEDSA L TRVKYSLT ...String: MKLSKDTTAL LKNFATINSG IMLKSGQFIM TRAVNGTTYA EANISDVIDF DVAIYDLNGF LGILSLVNDD AEISQSEDGN IKIADARST IFWPAADPST VVAPNKPIPF PVASAVTEIK AEDLQQLLRV SRGLQIDTIA ITVKEGKIVI NGFNKVEDSA L TRVKYSLT LGDYDGENTF NFIINMANMK MQPGNYKLLL WAKGKQGAAK FEGEHANYVV ALEADSTHDF UniProtKB:  Sliding clamp Sliding clamp |
-Macromolecule #4: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER / type: ligand / ID: 4 / Number of copies: 2 / Formula: AGS |
|---|---|
| Molecular weight | Theoretical: 523.247 Da |
| Chemical component information |  ChemComp-AGS: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 17937844 |
|---|---|
| Startup model | Type of model: INSILICO MODEL In silico model: 3D Ab-initio reconstruction from selected particles after 2D classification in cryoSPARC |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 3) / Software - details: ab-initio reconstruction |
| Final 3D classification | Number classes: 10 / Software - Name: cryoSPARC (ver. 3) / Software - details: heterogenous refinement |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 3) / Software - details: heterogenous refinement |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.21 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 3) / Software - details: Non-uniform refinement / Number images used: 455041 |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-8unh: |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X