-Search query
-Search result
Showing 1 - 50 of 746 items for (author: deng & h)
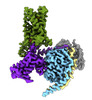
EMDB-43647: 
CryoEM structure of Gi-coupled TAS2R14 with cholesterol and an intracellular tastant
Method: single particle / : Kim Y, Gumpper RH, Roth BL
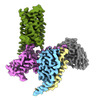
EMDB-43650: 
CryoEM structure of Ggust-coupled TAS2R14 with cholesterol and an intracellular tastant
Method: single particle / : Kim Y, Gumpper RH, Roth BL

EMDB-43656: 
CryoEM structure of Gi-coupled TAS2R14 with cholesterol and an intracellular tastant (Locally refined map)
Method: single particle / : Kim Y, Gumpper RH, Roth BL

EMDB-43657: 
CryoEM structure of Ggust-coupled TAS2R14 with cholesterol and an intracellular tastant (Locally refined map)
Method: single particle / : Kim Y, Gumpper RH, Roth BL

PDB-8vy7: 
CryoEM structure of Gi-coupled TAS2R14 with cholesterol and an intracellular tastant
Method: single particle / : Kim Y, Gumpper RH, Roth BL

PDB-8vy9: 
CryoEM structure of Ggust-coupled TAS2R14 with cholesterol and an intracellular tastant
Method: single particle / : Kim Y, Gumpper RH, Roth BL

EMDB-40650: 
Phosphoinositide phosphate 3 kinase gamma
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40651: 
Phosphoinositide phosphate 3 kinase gamma bound with ATP
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40652: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40653: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP and Gbetagamma
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40654: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP and two Gbetagamma subunits in State 1
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40655: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP and two Gbetagamma subunits in State 2
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

PDB-8so9: 
Phosphoinositide phosphate 3 kinase gamma
Method: single particle / : Chen CL, Tesmer JJG

PDB-8soa: 
Phosphoinositide phosphate 3 kinase gamma bound with ATP
Method: single particle / : Chen CL, Tesmer JJG

PDB-8sob: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP
Method: single particle / : Chen CL, Tesmer JJG

PDB-8soc: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP and Gbetagamma
Method: single particle / : Chen CL, Tesmer JJG

PDB-8sod: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP and two Gbetagamma subunits in State 1
Method: single particle / : Chen CL, Tesmer JJG

PDB-8soe: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP and two Gbetagamma subunits in State 2
Method: single particle / : Chen CL, Tesmer JJG

EMDB-38497: 
Cryo-EM structure of the ClpP degradation system in Streptomyces hawaiiensis
Method: single particle / : Xu X, Long F

EMDB-38535: 
Cryo-EM structure of the ClpC1:ClpP1P2 degradation complex in Streptomyces hawaiiensis
Method: single particle / : Xu X, Long F

EMDB-38536: 
Cryo-EM structure of the ClpC1:ClpP1P2 degradation complex in Streptomyces hawaiiensis
Method: single particle / : Xu X, Long F

EMDB-38537: 
Cryo-EM structure of ClpP1P2 in complex with ADEP1 from Streptomyces hawaiiensis
Method: single particle / : Xu X, Long F

PDB-8xn4: 
Cryo-EM structure of the ClpP degradation system in Streptomyces hawaiiensis
Method: single particle / : Xu X, Long F

PDB-8xon: 
Cryo-EM structure of the ClpC1:ClpP1P2 degradation complex in Streptomyces hawaiiensis
Method: single particle / : Xu X, Long F

PDB-8xoo: 
Cryo-EM structure of the ClpC1:ClpP1P2 degradation complex in Streptomyces hawaiiensis
Method: single particle / : Xu X, Long F

PDB-8xop: 
Cryo-EM structure of ClpP1P2 in complex with ADEP1 from Streptomyces hawaiiensis
Method: single particle / : Xu X, Long F

EMDB-43091: 
hGBP1 conformer on the bacterial outer membrane
Method: subtomogram averaging / : Zhu S, MacMicking J

EMDB-43153: 
hGBP1 conformer on the bacterial outer membrane
Method: subtomogram averaging / : Zhu S, MacMicking J

EMDB-35832: 
Cryo-EM structure of the GPR34 receptor in complex with the antagonist YL-365
Method: single particle / : Jia GW, Wang X, Zhang CB, Dong HH, Su ZM

PDB-8iyx: 
Cryo-EM structure of the GPR34 receptor in complex with the antagonist YL-365
Method: single particle / : Jia GW, Wang X, Zhang CB, Dong HH, Su ZM

EMDB-37516: 
BA.2(S375) Spike (S6P)/hACE2 complex
Method: single particle / : Wei X, Zhang Z

EMDB-37517: 
Local refinement of RBD-ACE2
Method: single particle / : Wei X, Zhang Z

EMDB-35461: 
Protomer 1 and 2 of the asymmetry trimer of the Cul2-Rbx1-EloBC-FEM1B ubiquitin ligase complex
Method: single particle / : Dai Z, Liang L, Yin YX

EMDB-36182: 
An asymmetry dimer of the Cul2-Rbx1-EloBC-FEM1B ubiquitin ligase complexed with BEX2
Method: single particle / : Dai Z, Liang L, Yin YX

EMDB-36183: 
Cryo-EM structure of neddylated Cul2-Rbx1-EloBC-FEM1B complexed with FNIP1-FLCN
Method: single particle / : Dai Z, Liang L, Yin YX

PDB-8ij1: 
Protomer 1 and 2 of the asymmetry trimer of the Cul2-Rbx1-EloBC-FEM1B ubiquitin ligase complex
Method: single particle / : Dai Z, Liang L, Yin YX

PDB-8je1: 
An asymmetry dimer of the Cul2-Rbx1-EloBC-FEM1B ubiquitin ligase complexed with BEX2
Method: single particle / : Dai Z, Liang L, Yin YX

PDB-8je2: 
Cryo-EM structure of neddylated Cul2-Rbx1-EloBC-FEM1B complexed with FNIP1-FLCN
Method: single particle / : Dai Z, Liang L, Yin YX

EMDB-36369: 
Focused map on the aCTD-AfsR(T337A) region of the Streptomyces coelicolor RNAP-promoter open complex with AfsR(T337A) dimer
Method: single particle / : Wang Y, Zheng J

EMDB-36370: 
AfsR(T337A) transcription activation complex
Method: single particle / : Wang Y, Zheng J

EMDB-43542: 
ELIC5 with cysteamine in 2:1:1 POPC:POPE:POPG nanodisc in open conformation
Method: single particle / : Petroff II JT, Deng Z, Rau MJ, Fitzpatrick JAJ, Yuan P, Cheng WWL

PDB-8vuw: 
ELIC5 with cysteamine in 2:1:1 POPC:POPE:POPG nanodisc in open conformation
Method: single particle / : Petroff II JT, Deng Z, Rau MJ, Fitzpatrick JAJ, Yuan P, Cheng WWL

EMDB-38543: 
Cryo-EM map of the internal RuBisCOs in the alpha-carboxysome from Prochlorococcus MED4
Method: single particle / : Jiang YL, Zhou RQ, Zhou CZ, Zeng QL

EMDB-38544: 
Cryo-EM map of the intact shell of alpha-carboxysome from Prochlorococcus MED4
Method: single particle / : Jiang YL, Zhou RQ, Zhou CZ, Zeng QL

EMDB-37902: 
Cryo-EM structure of the alpha-carboxysome shell vertex from Prochlorococcus MED4
Method: single particle / : Jiang YL, Zhou RQ, Zhou CZ, Zeng QL

EMDB-36368: 
Cryo-EM structure of CCHFV envelope protein Gc trimer in complex with Gc13 Fab
Method: single particle / : Chong T, Cao S

EMDB-36406: 
CCHFV envelope protein Gc in complex with Gc8
Method: single particle / : Chong T, Cao S
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search






 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
