[English] 日本語
 Yorodumi
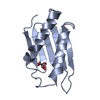
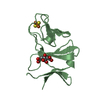
Yorodumi- PDB-3a2e: Crystal structure of ginkbilobin-2, the novel antifungal protein ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3a2e | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of ginkbilobin-2, the novel antifungal protein from Ginkgo biloba seeds | |||||||||
 Components Components | Ginkbilobin-2 | |||||||||
 Keywords Keywords | PLANT PROTEIN / DOMAIN 26 UNKNOWN FUNCTION (DUF26) / C-X8-C-X2-C MOTIF / ANTIFUNGAL PROTEIN / EMBRYO-ABUNDANT PROTEIN (EAP) | |||||||||
| Function / homology |  Function and homology information Function and homology informationinduction of programmed cell death / aspartic-type endopeptidase inhibitor activity / D-mannose binding / defense response to fungus / actin binding / killing of cells of another organism / defense response to bacterium / extracellular space Similarity search - Function | |||||||||
| Biological species |  Ginkgo biloba (maidenhair tree) Ginkgo biloba (maidenhair tree) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MAD / Resolution: 2.38 Å MAD / Resolution: 2.38 Å | |||||||||
 Authors Authors | Miyakawa, T. / Miyazono, K. / Sawano, Y. / Hatano, K. / Tanokura, M. | |||||||||
 Citation Citation |  Journal: Proteins / Year: 2009 Journal: Proteins / Year: 2009Title: Crystal structure of ginkbilobin-2 with homology to the extracellular domain of plant cysteine-rich receptor-like kinases Authors: Miyakawa, T. / Miyazono, K. / Sawano, Y. / Hatano, K. / Tanokura, M. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3a2e.cif.gz 3a2e.cif.gz | 101 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3a2e.ent.gz pdb3a2e.ent.gz | 78.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3a2e.json.gz 3a2e.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/a2/3a2e https://data.pdbj.org/pub/pdb/validation_reports/a2/3a2e ftp://data.pdbj.org/pub/pdb/validation_reports/a2/3a2e ftp://data.pdbj.org/pub/pdb/validation_reports/a2/3a2e | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||
| 2 | 
| ||||||||||||
| 3 | 
| ||||||||||||
| 4 | 
| ||||||||||||
| Unit cell |
| ||||||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 11671.059 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Ginkgo biloba (maidenhair tree) / Plasmid: pET26b / Production host: Ginkgo biloba (maidenhair tree) / Plasmid: pET26b / Production host:  #2: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 2 X-RAY DIFFRACTION / Number of used crystals: 2 |
|---|
- Sample preparation
Sample preparation
| Crystal |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow |
|
-Data collection
| Diffraction |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source |
| |||||||||||||||
| Detector |
| |||||||||||||||
| Radiation |
| |||||||||||||||
| Radiation wavelength |
| |||||||||||||||
| Reflection twin | Operator: l,-k,h / Fraction: 0.485 | |||||||||||||||
| Reflection | Resolution: 2.38→50 Å / Num. obs: 39529 / % possible obs: 99.7 % / Redundancy: 15.2 % / Rsym value: 0.061 / Net I/σ(I): 61.2 / Num. measured all: 602150 | |||||||||||||||
| Reflection shell | Resolution: 2.38→2.47 Å / Redundancy: 14.5 % / Mean I/σ(I) obs: 8.2 / Rsym value: 0.375 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MAD / Resolution: 2.38→43.17 Å / Occupancy max: 1 / Occupancy min: 0.98 / Cross valid method: THROUGHOUT / σ(F): 1.36 / Phase error: 21.09 / Stereochemistry target values: TWIN_LSQ_F MAD / Resolution: 2.38→43.17 Å / Occupancy max: 1 / Occupancy min: 0.98 / Cross valid method: THROUGHOUT / σ(F): 1.36 / Phase error: 21.09 / Stereochemistry target values: TWIN_LSQ_F
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 87.147 Å2 / ksol: 0.346 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 70.92 Å2 / Biso mean: 34.8 Å2 / Biso min: 25.03 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.38→43.17 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 20 / % reflection obs: 98 %
|
 Movie
Movie Controller
Controller









 PDBj
PDBj

