[English] 日本語
 Yorodumi
Yorodumi- PDB-2k0p: Determination of a Protein Structure in the Solid State from NMR ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2k0p | ||||||
|---|---|---|---|---|---|---|---|
| Title | Determination of a Protein Structure in the Solid State from NMR Chemical Shifts | ||||||
 Components Components | Immunoglobulin G-binding protein G | ||||||
 Keywords Keywords | PROTEIN BINDING / solid-state / chemical shift restraints / GB1 / Cell wall / IgG-binding protein / Peptidoglycan-anchor / Secreted | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  Streptococcus sp. group G (bacteria) Streptococcus sp. group G (bacteria) | ||||||
| Method | SOLID-STATE NMR / MD, MC hybrid refinment against Target Fuction weighted by chemical shift accuracy, a molecular mechanics force field | ||||||
 Authors Authors | Robustelli, P. / Cavalli, A. / Salvatella, X. / Vendruscolo, M. | ||||||
 Citation Citation |  Journal: Structure / Year: 2008 Journal: Structure / Year: 2008Title: Determination of protein structures in the solid state from NMR chemical shifts. Authors: Robustelli, P. / Cavalli, A. / Vendruscolo, M. #1:  Journal: Angew.Chem.Int.Ed.Engl. / Year: 2007 Journal: Angew.Chem.Int.Ed.Engl. / Year: 2007Title: Solid-state protein-structure determination with proton-detected triple-resonance 3D magic-angle-spinning NMR spectroscopy. Authors: Zhou, D.H. / Shea, J.J. / Nieuwkoop, A.J. / Franks, W.T. / Wylie, B.J. / Mullen, C. / Sandoz, D. / Rienstra, C.M. #2: Journal: J.Am.Chem.Soc. / Year: 2007 Title: Proton-detected solid-state NMR spectroscopy of fully protonated proteins at 40 kHz magic-angle spinning. Authors: Zhou, D.H. / Shah, G. / Cormos, M. / Mullen, C. / Sandoz, D. / Rienstra, C.M. #3: Journal: Proc.Natl.Acad.Sci.Usa / Year: 2007 Title: Protein structure determination from NMR chemical shifts. Authors: Cavalli, A. / Salvatella, X. / Dobson, C.M. / Vendruscolo, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2k0p.cif.gz 2k0p.cif.gz | 20.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2k0p.ent.gz pdb2k0p.ent.gz | 12.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2k0p.json.gz 2k0p.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/k0/2k0p https://data.pdbj.org/pub/pdb/validation_reports/k0/2k0p ftp://data.pdbj.org/pub/pdb/validation_reports/k0/2k0p ftp://data.pdbj.org/pub/pdb/validation_reports/k0/2k0p | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 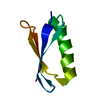
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Antibody | Mass: 6228.809 Da / Num. of mol.: 1 / Fragment: GB1 / Mutation: T2Q Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Streptococcus sp. group G (bacteria) / Gene: spg / Production host: Streptococcus sp. group G (bacteria) / Gene: spg / Production host:  |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLID-STATE NMR Method details: Solid-state structure of Beta-1 Immunoglobulin binding domain of protein G (GB1) solved from Ha, Ca, Cb, and N backbone chemical shifts. | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||||||||||
| NMR details | Text: For Experimental Data See BMRB Entry 15156 |
- Sample preparation
Sample preparation
| Details | Contents: 10 mg/mL [U-100% 13C; U-100% 15N] GB1, 0.5 v/v Methyl Pentane diol, 0.25 v/v Isopropanol Solvent system: 0.5 v/v Methyl Pentane diol/0.25 v/v Isopropanol | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||
| Sample conditions | Pressure: 1 atm / Temperature: 278 K |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: MD, MC hybrid refinment against Target Fuction weighted by chemical shift accuracy, a molecular mechanics force field Software ordinal: 1 Details: Structure Selection and Refinement were performed according to CHESHIRE protocol for calculation of structures from NMR Chemical Shifts (Cavalli et al., 2007, PNAS, 104, 9615-9620) | ||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||
| NMR ensemble | Conformer selection criteria: target function / Conformers calculated total number: 1500 / Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller









 PDBj
PDBj

