[English] 日本語
 Yorodumi
Yorodumi- PDB-272d: PARALLEL AND ANTIPARALLEL (G.GC)2 TRIPLE HELIX FRAGMENTS IN A CRY... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 272d | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | PARALLEL AND ANTIPARALLEL (G.GC)2 TRIPLE HELIX FRAGMENTS IN A CRYSTAL STRUCTURE | ||||||||||||||||||||
 Components Components | DNA (5'-D(* Keywords KeywordsDNA / B-DNA / DOUBLE HELIX / INTERMOLECULAR BASE TRIPLET / OVERHANGING BASES | Function / homology | DNA |  Function and homology information Function and homology informationMethod |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2 Å MOLECULAR REPLACEMENT / Resolution: 2 Å  Authors AuthorsVlieghe, D. / Van Meervelt, L. / Dautant, A. / Gallois, B. / Precigoux, G. / Kennard, O. |  Citation Citation Journal: Science / Year: 1996 Journal: Science / Year: 1996Title: Parallel and antiparallel (G.GC)2 triple helix fragments in a crystal structure. Authors: Vlieghe, D. / Van Meervelt, L. / Dautant, A. / Gallois, B. / Precigoux, G. / Kennard, O. History |
Remark 300 | BIOMOLECULE: 1 THIS ENTRY CONTAINS THE CRYSTALLOGRAPHIC ASYMMETRIC UNIT WHICH CONSISTS OF 2 ... BIOMOLECULE: 1 THIS ENTRY CONTAINS THE CRYSTALLOGRAPHIC ASYMMETRIC UNIT WHICH CONSISTS OF 2 CHAIN(S). SEE REMARK 350 FOR INFORMATION ON GENERATING THE BIOLOGICAL MOLECULE(S). TWO TYPES OF TRIPLEXES CAN BE GENERATED - PARALLEL TRIPLEX AND ANTIPARALLEL TRIPLEX. ONLY THE SYMMETRY OPERATIONS WHICH GENERATE THE PARALLEL TRIPLEX ARE SHOWN IN REMARK 350. THE PARALLEL TRIPLEX IS GENERATED BY APPLYING SYMMETRY OPERATION 1/2-X, 1-Y, 1/2+Z. THE ANTIPARALLEL TRIPLEX IS GENERATED BY APPLYING SYMMETRY OPERATION 1-X, 1/2+Y, 1/2-Z. | |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  272d.cif.gz 272d.cif.gz | 20.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb272d.ent.gz pdb272d.ent.gz | 12.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  272d.json.gz 272d.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/72/272d https://data.pdbj.org/pub/pdb/validation_reports/72/272d ftp://data.pdbj.org/pub/pdb/validation_reports/72/272d ftp://data.pdbj.org/pub/pdb/validation_reports/72/272d | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 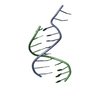
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 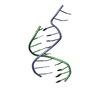
| ||||||||
| Unit cell |
|
- Components
Components
| #1: DNA chain | Mass: 3085.029 Da / Num. of mol.: 2 / Source method: obtained synthetically #2: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2 Å3/Da / Density % sol: 38.55 % | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 289 K / Method: vapor diffusion / pH: 6 / Details: pH 6.00, VAPOR DIFFUSION, temperature 289.00K | ||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions |
| ||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 16 ℃ / pH: 6 | ||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Ambient temp details: ROOM TEMPERATURE |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: LURE SYNCHROTRON / Site: LURE  / Beamline: D41A / Beamline: D41A |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Jun 1, 1995 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Relative weight: 1 |
| Reflection | Resolution: 2→14.5 Å / Num. all: 12303 / Num. obs: 3534 / % possible obs: 94.6 % / Redundancy: 3 % / Rsym value: 0.046 / Net I/σ(I): 5.4 |
| Reflection shell | Resolution: 2→2.1 Å / Redundancy: 2.4 % / Mean I/σ(I) obs: 3.8 / Rsym value: 0.193 / % possible all: 86.3 |
| Reflection | *PLUS Highest resolution: 2 Å / Lowest resolution: 14.5 Å / % possible obs: 94.6 % / Redundancy: 3 % / Num. measured all: 12303 / Rmerge(I) obs: 0.046 |
| Reflection shell | *PLUS Highest resolution: 2 Å / Lowest resolution: 2.1 Å / % possible obs: 86.3 % / Redundancy: 2.4 % / Rmerge(I) obs: 0.193 / Mean I/σ(I) obs: 3.8 |
- Processing
Processing
| Software |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 2→10 Å / Num. parameters: 1818 / Num. restraintsaints: 1990 / σ(F): 0 / MOLECULAR REPLACEMENT / Resolution: 2→10 Å / Num. parameters: 1818 / Num. restraintsaints: 1990 / σ(F): 0 /
| |||||||||||||||
| Solvent computation | Solvent model: BABINET'S PRINCIPLE | |||||||||||||||
| Displacement parameters | Biso mean: 30.6 Å2 | |||||||||||||||
| Refine Biso |
| |||||||||||||||
| Refine analyze | Num. disordered residues: 0 | |||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→10 Å
| |||||||||||||||
| Software | *PLUS Name: SHELXL-93 / Classification: refinement | |||||||||||||||
| Refinement | *PLUS Highest resolution: 2 Å / Lowest resolution: 10 Å / Num. reflection obs: 3212 / σ(F): 0 | |||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||
| Displacement parameters | *PLUS Biso mean: 30.6 Å2 |
 Movie
Movie Controller
Controller









 PDBj
PDBj


