[English] 日本語
 Yorodumi
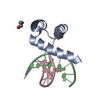
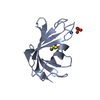
Yorodumi- PDB-1gmm: Carbohydrate binding module CBM6 from xylanase U Clostridium ther... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1gmm | ||||||
|---|---|---|---|---|---|---|---|
| Title | Carbohydrate binding module CBM6 from xylanase U Clostridium thermocellum | ||||||
 Components Components | CBM6 | ||||||
 Keywords Keywords | XYLANASE / CARBOHYDRATE BINDING MODULE / CBM FAMILY 6 / XYLAN BINDING | ||||||
| Function / homology |  Function and homology information Function and homology informationhydrolase activity, acting on carbon-nitrogen (but not peptide) bonds / endo-1,4-beta-xylanase activity / endo-1,4-beta-xylanase / xylan catabolic process / carbohydrate binding / metal ion binding Similarity search - Function | ||||||
| Biological species |  CLOSTRIDIUM THERMOCELLUM (bacteria) CLOSTRIDIUM THERMOCELLUM (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MAD / Resolution: 2 Å MAD / Resolution: 2 Å | ||||||
 Authors Authors | Czjzek, M. / Mosbah, A. / Bolam, D. / Allouch, J. / Zamboni, V. / Henrissat, B. / Gilbert, H.J. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2001 Journal: J.Biol.Chem. / Year: 2001Title: The Location of the Ligand-Binding Site of Carbohydrate-Binding Modules that Have Evolved from a Common Sequence is not Conserved. Authors: Czjzek, M. / Bolam, D. / Mosbah, A. / Allouch, J. / Fontes, C.M. / Ferreira, L.M. / Bornet, O. / Zamboni, V. / Darbon, H. / Smith, N.L. / Black, G.W. / Henrissat, B. / Gilbert, H.J. #1: Journal: Biochem.J. / Year: 1999 Title: Homologous Xylanases from Clostridium Thermocellum: Evidence for Bi-Functional Activity, Synergism between Xylanase Catalytic Modules and the Presence of Xylan-Binding Domains in Enzyme Complexes Authors: Fernandes, A. / Fontes, C.M.G.A. / Gilbert, H.J. / Hazelwood, G.P. / Fernandes, T.H. / Ferreira, L.M.A. / Gilbert, H.J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1gmm.cif.gz 1gmm.cif.gz | 42.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1gmm.ent.gz pdb1gmm.ent.gz | 29.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1gmm.json.gz 1gmm.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/gm/1gmm https://data.pdbj.org/pub/pdb/validation_reports/gm/1gmm ftp://data.pdbj.org/pub/pdb/validation_reports/gm/1gmm ftp://data.pdbj.org/pub/pdb/validation_reports/gm/1gmm | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 14115.347 Da / Num. of mol.: 1 / Fragment: XYLAN BINDING MODULE (DOMAIN), RESIDUE 248-380 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  CLOSTRIDIUM THERMOCELLUM (bacteria) / Strain: F1 / Production host: CLOSTRIDIUM THERMOCELLUM (bacteria) / Strain: F1 / Production host:  |
|---|---|
| #2: Chemical | ChemComp-SO4 / |
| #3: Chemical | ChemComp-NA / |
| #4: Chemical | ChemComp-CA / |
| #5: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.73 Å3/Da / Density % sol: 55 % | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 5.5 Details: 30% PEG 4000, 100MM NA-CITRATE PH 5.5, 100MM (NH4)2SO4 | ||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 4 ℃ / pH: 5 / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID14-4 / Wavelength: 0.9793, 0.9795, 0.9465, 0.933 / Beamline: ID14-4 / Wavelength: 0.9793, 0.9795, 0.9465, 0.933 | |||||||||||||||
| Detector | Detector: CCD / Date: Dec 15, 2000 / Details: MIRRORS | |||||||||||||||
| Radiation | Protocol: MAD / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||
| Radiation wavelength |
| |||||||||||||||
| Reflection | Resolution: 2→39.22 Å / Num. obs: 11981 / % possible obs: 99.3 % / Observed criterion σ(I): 0.01 / Redundancy: 6.6 % / Rmerge(I) obs: 0.05 / Net I/σ(I): 9.3 | |||||||||||||||
| Reflection shell | Resolution: 2→2.11 Å / Redundancy: 6.2 % / Rmerge(I) obs: 0.204 / Mean I/σ(I) obs: 1.2 / % possible all: 98.2 | |||||||||||||||
| Reflection | *PLUS Rmerge(I) obs: 0.055 | |||||||||||||||
| Reflection shell | *PLUS % possible obs: 98.2 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MAD / Resolution: 2→39 Å / SU B: 3.345 / SU ML: 0.095 / Cross valid method: THROUGHOUT / σ(F): 0.01 / ESU R Free: 0.142 MAD / Resolution: 2→39 Å / SU B: 3.345 / SU ML: 0.095 / Cross valid method: THROUGHOUT / σ(F): 0.01 / ESU R Free: 0.142 Details: TLS REFINEMENT WAS PERFORMED ON 6 GROUPS. FOUR RESIDUES, THR A 65, THR A 67, SER A 99 AND THR A 103 HAVE BEEN MODELED WITH MULTIPLE ALTERNATE CONFORMATIONS.
| ||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→39 Å
| ||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 2.1 Å / Lowest resolution: 32 Å / Rfactor obs: 0.2 / Rfactor Rfree: 0.216 / Rfactor Rwork: 0.2 | ||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller










 PDBj
PDBj








