+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1g7z | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
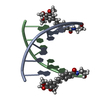
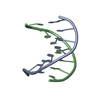
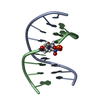
| Title | NMR SOLUTION STRUCTURE OF D(CGCTAGCG)2 | ||||||||||||||||||
 Components Components | 5'-D(* Keywords Keywords DNA / TOTO / C13 DYNAMICS / CONFORMATIONAL EXCHANGE / PHOSPHATE CONFORMATION / DEOXYRIBOSE CONFORMATION / HELICAL PARAMETER / DNA / TOTO / C13 DYNAMICS / CONFORMATIONAL EXCHANGE / PHOSPHATE CONFORMATION / DEOXYRIBOSE CONFORMATION / HELICAL PARAMETER /  ORDER PARAMETER ORDER PARAMETERFunction / homology |  DNA DNA Function and homology information Function and homology informationMethod |  SOLUTION NMR / The RANDMARDI procedure of the complete relaxation matrix analysis method, MARDIGRAS, was used to calculate interproton distance bounds from the integrated NOESY cross-peak intensities. These distance bounds were then used as restraints in an RMD procedure to yield 20 structures. SOLUTION NMR / The RANDMARDI procedure of the complete relaxation matrix analysis method, MARDIGRAS, was used to calculate interproton distance bounds from the integrated NOESY cross-peak intensities. These distance bounds were then used as restraints in an RMD procedure to yield 20 structures.  Authors AuthorsIsaacs, R.J. / Spielmann, H.P. |  Citation Citation Journal: J.Mol.Biol. / Year: 2001 Journal: J.Mol.Biol. / Year: 2001Title: Relationship of DNA structure to internal dynamics: correlation of helical parameters from NOE-based NMR solution structures of d(GCGTACGC)(2) and d(CGCTAGCG)(2) with (13)C order parameters ...Title: Relationship of DNA structure to internal dynamics: correlation of helical parameters from NOE-based NMR solution structures of d(GCGTACGC)(2) and d(CGCTAGCG)(2) with (13)C order parameters implies conformational coupling in dinucleotide units. Authors: Isaacs, R.J. / Spielmann, H.P. #1:  Journal: Biochemistry / Year: 1998 Journal: Biochemistry / Year: 1998Title: Dynamics of a Bis-intercalator DNA Complex by 1H-Detected Natural Abundance 13C NMR Spectroscopy Authors: Spielmann, H.P. #2:  Journal: Biochemistry / Year: 1995 Journal: Biochemistry / Year: 1995Title: Solution structure of a DNA complex with the fluorescent bis-intercalator TOTO determined by NMR spectroscopy. Authors: Spielmann, H.P. / Wemmer, D.E. / Jacobsen, J.P. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1g7z.cif.gz 1g7z.cif.gz | 199.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1g7z.ent.gz pdb1g7z.ent.gz | 172.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1g7z.json.gz 1g7z.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/g7/1g7z https://data.pdbj.org/pub/pdb/validation_reports/g7/1g7z ftp://data.pdbj.org/pub/pdb/validation_reports/g7/1g7z ftp://data.pdbj.org/pub/pdb/validation_reports/g7/1g7z | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 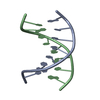
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 2427.605 Da / Num. of mol.: 2 / Source method: obtained synthetically / Details: phosphoramadites on solid support |
|---|
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR |
|---|---|
| NMR experiment | Type : 2D : 2D  NOESY NOESY |
| NMR details | Text: This structure was determined using standard 2D homonuclear techniques. |
- Sample preparation
Sample preparation
| Details | Contents: 4 mM DNA duplex / Solvent system: 99.96% D2O |
|---|---|
| Sample conditions | Ionic strength: NaCl(100mM),PO4-(20mM),NaN3(10mM),EDTA(0.1mM) pH: 7.0 / Pressure: ambient / Temperature: 298 K |
Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Varian INOVA / Manufacturer: Varian / Model : INOVA / Field strength: 500 MHz : INOVA / Field strength: 500 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: The RANDMARDI procedure of the complete relaxation matrix analysis method, MARDIGRAS, was used to calculate interproton distance bounds from the integrated NOESY cross-peak intensities. These ...Method: The RANDMARDI procedure of the complete relaxation matrix analysis method, MARDIGRAS, was used to calculate interproton distance bounds from the integrated NOESY cross-peak intensities. These distance bounds were then used as restraints in an RMD procedure to yield 20 structures. Software ordinal: 1 Details: The structures are based on a total of 540 restraints, 518 are NOE-derived distance constraints and 22 distance restraints from hydrogen bonds. | ||||||||||||||||||||||||
| NMR representative | Selection criteria: lowest pairwise rmsd from other conformers | ||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: back calculated data agree with experimental NOESY spectrum Conformers calculated total number: 20 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller








 PDBj
PDBj

