[English] 日本語
 Yorodumi
Yorodumi- PDB-1g2b: ALPHA-SPECTRIN SRC HOMOLOGY 3 DOMAIN, CIRCULAR PERMUTANT, CUT AT ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1g2b | ||||||
|---|---|---|---|---|---|---|---|
| Title | ALPHA-SPECTRIN SRC HOMOLOGY 3 DOMAIN, CIRCULAR PERMUTANT, CUT AT N47-D48 | ||||||
 Components Components | SPECTRIN ALPHA CHAIN | ||||||
 Keywords Keywords | METAL BINDING PROTEIN / CAPPING PROTEIN / CALCIUM-BINDING / DUPLICATION / SH3 DOMAIN / CYTOSKELETON | ||||||
| Function / homology |  Function and homology information Function and homology informationcostamere / actin filament capping / cortical actin cytoskeleton / cell projection / actin filament binding / cell junction / actin cytoskeleton organization / calmodulin binding / calcium ion binding / plasma membrane Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / AB INITIO / Resolution: 1.12 Å SYNCHROTRON / AB INITIO / Resolution: 1.12 Å | ||||||
 Authors Authors | Berisio, R. / Viguera, A.R. / Serrano, L. / Wilmanns, M. | ||||||
 Citation Citation |  Journal: Acta Crystallogr.,Sect.D / Year: 2001 Journal: Acta Crystallogr.,Sect.D / Year: 2001Title: Atomic resolution structure of a mutant of the spectrin SH3 domain. Authors: Berisio, R. / Viguera, A. / Serrano, L. / Wilmanns, M. #1:  Journal: Biochemistry / Year: 1999 Journal: Biochemistry / Year: 1999Title: Thermodynamic Analysis of alpha-spectrin SH3 and Two of Its Circular Permutants with Different Loop Lengths: Discerning the Reasons for Rapid Folding in Proteins Authors: Martinez, J.C. / Viguera, A.R. / Berisio, R. / Wilmanns, M. / Mateo, P.L. / Filimonov, V.V. / Serrano, L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1g2b.cif.gz 1g2b.cif.gz | 46.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1g2b.ent.gz pdb1g2b.ent.gz | 32.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1g2b.json.gz 1g2b.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/g2/1g2b https://data.pdbj.org/pub/pdb/validation_reports/g2/1g2b ftp://data.pdbj.org/pub/pdb/validation_reports/g2/1g2b ftp://data.pdbj.org/pub/pdb/validation_reports/g2/1g2b | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 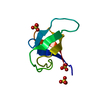
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 7129.172 Da / Num. of mol.: 1 / Fragment: SH3 DOMAIN Source method: isolated from a genetically manipulated source Details: CIRCULAR PERMUTANT, CUT AT N47-D48, INS(M-D48), INS(SG-T4) Source: (gene. exp.)   | ||
|---|---|---|---|
| #2: Chemical | | #3: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.49 Å3/Da / Density % sol: 50.51 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / Details: VAPOR DIFFUSION, HANGING DROP, temperature 298K |
| Crystal grow | *PLUS Details: Viguera, A.R., (1996) Nature Struct. Biol., 3, 874. |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  EMBL/DESY, HAMBURG EMBL/DESY, HAMBURG  / Beamline: X11 / Beamline: X11 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Relative weight: 1 |
| Reflection | Resolution: 1.12→14 Å / Num. all: 24461 / Num. obs: 24461 / % possible obs: 93.4 % / Rmerge(I) obs: 0.071 |
| Reflection shell | Resolution: 1.12→1.2 Å / Rmerge(I) obs: 0.14 / % possible all: 91.2 |
| Reflection | *PLUS Num. measured all: 102998 |
| Reflection shell | *PLUS % possible obs: 91.2 % |
- Processing
Processing
| Software |
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: AB INITIO / Resolution: 1.12→14 Å / Num. parameters: 634 / Num. restraintsaints: 742 / Cross valid method: R-FREE / Stereochemistry target values: ENGH AND HUBER
| ||||||||||||||||
| Solvent computation | Solvent model: MOEWS & KRETSINGER, J.MOL.BIOL.91(1973)201-228 | ||||||||||||||||
| Refine analyze | Occupancy sum hydrogen: 442 / Occupancy sum non hydrogen: 664 | ||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.12→14 Å
| ||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||
| LS refinement shell | Resolution: 1.12→14 Å
| ||||||||||||||||
| Software | *PLUS Name: SHELXL-97 / Classification: refinement | ||||||||||||||||
| Refinement | *PLUS σ(F): 4 / % reflection Rfree: 5 % / Rfactor all: 0.148 / Rfactor obs: 0.136 | ||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller









 PDBj
PDBj


