+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1bg8 | ||||||
|---|---|---|---|---|---|---|---|
| Title | HDEA FROM ESCHERICHIA COLI | ||||||
 Components Components | HDEA | ||||||
 Keywords Keywords | PERIPLASMIC / HDEA | ||||||
| Function / homology |  Function and homology information Function and homology informationcellular stress response to acidic pH / cellular response to acidic pH / protein folding chaperone / unfolded protein binding / protein-folding chaperone binding / protein folding / outer membrane-bounded periplasmic space / protein homodimerization activity / identical protein binding Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MIR / Resolution: 2.2 Å MIR / Resolution: 2.2 Å | ||||||
 Authors Authors | Yang, F. / Gustafson, K.R. / Boyd, M.R. / Wlodawer, A. | ||||||
 Citation Citation |  Journal: Nat.Struct.Biol. / Year: 1998 Journal: Nat.Struct.Biol. / Year: 1998Title: Crystal structure of Escherichia coli HdeA. Authors: Yang, F. / Gustafson, K.R. / Boyd, M.R. / Wlodawer, A. | ||||||
| History |
|
- Structure visualization
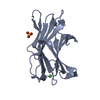
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1bg8.cif.gz 1bg8.cif.gz | 57.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1bg8.ent.gz pdb1bg8.ent.gz | 43.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1bg8.json.gz 1bg8.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bg/1bg8 https://data.pdbj.org/pub/pdb/validation_reports/bg/1bg8 ftp://data.pdbj.org/pub/pdb/validation_reports/bg/1bg8 ftp://data.pdbj.org/pub/pdb/validation_reports/bg/1bg8 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| |||||||||||||||||||||
| 2 | 
| |||||||||||||||||||||
| Unit cell |
| |||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS oper:
|
- Components
Components
| #1: Protein | Mass: 9752.882 Da / Num. of mol.: 3 / Source method: isolated from a natural source / Source: (natural)  #2: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.2 Å3/Da / Density % sol: 45 % | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 3.6 Details: 36% PEG400, 5% GLYCEROL, 50MM SODIUM CITRATE, PH 3.6 | ||||||||||||||||||||
| Crystal | *PLUS | ||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop | ||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 293 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 |
| Detector | Type: MAR scanner 345 mm plate / Detector: IMAGE PLATE / Date: Feb 16, 1998 / Details: MIRRORS |
| Radiation | Monochromator: DOUBLE CRYSTAL SI(111) / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.2→40 Å / Num. obs: 12863 / % possible obs: 97.2 % / Observed criterion σ(I): -3 / Redundancy: 3.6 % / Rsym value: 0.071 / Net I/σ(I): 8.4 |
| Reflection shell | Resolution: 2.2→2.26 Å / Rsym value: 0.473 / % possible all: 94.8 |
| Reflection | *PLUS Rmerge(I) obs: 0.071 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MIR / Resolution: 2.2→10 Å / Data cutoff high absF: 100000 / Data cutoff low absF: 0.1 / Cross valid method: FREE R / σ(F): 2 MIR / Resolution: 2.2→10 Å / Data cutoff high absF: 100000 / Data cutoff low absF: 0.1 / Cross valid method: FREE R / σ(F): 2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 40.3 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.2→10 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.2→2.3 Å / Total num. of bins used: 8
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.1 / Classification: refinement X-PLOR / Version: 3.1 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller












 PDBj
PDBj
