[English] 日本語
 Yorodumi
Yorodumi- SASDCZ2: Yeast CTDsome: transcription termination factor Rtt103p/C-termina... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDCZ2 |
|---|---|
 Sample Sample | Yeast CTDsome: transcription termination factor Rtt103p/C-terminal domain of DNA-directed RNA polymerase II subunit RPB1 (Rtt103 1-246-phospho-mimetic complex)
|
| Function / homology |  Function and homology information Function and homology informationRNA polymerase II C-terminal domain phosphoserine binding / transposable element silencing / mRNA 3'-end processing / Processing of Capped Intron-Containing Pre-mRNA / RNA Pol II CTD phosphorylation and interaction with CE / Formation of the Early Elongation Complex / mRNA Capping / RNA polymerase II transcribes snRNA genes / TP53 Regulates Transcription of DNA Repair Genes / RNA Polymerase II Promoter Escape ...RNA polymerase II C-terminal domain phosphoserine binding / transposable element silencing / mRNA 3'-end processing / Processing of Capped Intron-Containing Pre-mRNA / RNA Pol II CTD phosphorylation and interaction with CE / Formation of the Early Elongation Complex / mRNA Capping / RNA polymerase II transcribes snRNA genes / TP53 Regulates Transcription of DNA Repair Genes / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance / RNA Polymerase II Pre-transcription Events / RNA-templated transcription / Formation of TC-NER Pre-Incision Complex / Gap-filling DNA repair synthesis and ligation in TC-NER / RNA polymerase II complex binding / Estrogen-dependent gene expression / Dual incision in TC-NER / translesion synthesis / RNA polymerase II, core complex / transcription initiation at RNA polymerase II promoter / transcription elongation by RNA polymerase II / cytoplasmic stress granule / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / site of double-strand break / transcription by RNA polymerase II / chromatin / mitochondrion / DNA binding / metal ion binding / nucleus Similarity search - Function |
| Biological species |  |
 Citation Citation |  Date: 2017 Oct 4 Date: 2017 Oct 4Title: Structure and dynamics of the RNAPII CTDsome with Rtt103 Authors: Jasnovidova O / Klumpler T / Kubicek K / Kalynych S / Plevka P |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDCZ2 SASDCZ2 |
|---|
-Related structure data
| Similar structure data |
|---|
- External links
External links
| Related items in Molecule of the Month |
|---|
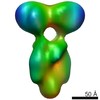
-Models
| Model #1182 |  Type: mix / Software: (05) / Radius of dummy atoms: 1.90 A / Chi-square value: 0.79 / P-value: 0.545500  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|---|
| Model #1195 |  Type: mix / Software: (05) / Radius of dummy atoms: 1.90 A / Chi-square value: 0.81 / P-value: 0.966400  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
| Model #1197 |  Type: mix / Software: (05) / Radius of dummy atoms: 1.90 A / Chi-square value: 0.81 / P-value: 0.321000  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
- Sample
Sample
 Sample Sample | Name: Yeast CTDsome: transcription termination factor Rtt103p/C-terminal domain of DNA-directed RNA polymerase II subunit RPB1 (Rtt103 1-246-phospho-mimetic complex) Specimen concentration: 1 mg/ml / Entity id: 615 / 616 |
|---|---|
| Buffer | Name: 25 mM KH2PO4, 300 mM KCl, 10 mM BME / pH: 6.5 |
| Entity #615 | Type: protein / Description: Regulator of Ty1 transposition protein 103 / Formula weight: 28.627 / Num. of mol.: 2 Source: Saccharomyces cerevisiae (strain ATCC 204508 / S288c) References: UniProt: Q05543 Sequence: MPFSSEQFTT KLNTLEDSQE SISSASKWLL LQYRDAPKVA EMWKEYMLRP SVNTRRKLLG LYLMNHVVQQ AKGQKIIQFQ DSFGKVAAEV LGRINQEFPR DLKKKLSRVV NILKERNIFS KQVVNDIERS LKTESSPVEA LVLPQKLKDF AKDYEKLVKM HHNVCAMKMR ...Sequence: MPFSSEQFTT KLNTLEDSQE SISSASKWLL LQYRDAPKVA EMWKEYMLRP SVNTRRKLLG LYLMNHVVQQ AKGQKIIQFQ DSFGKVAAEV LGRINQEFPR DLKKKLSRVV NILKERNIFS KQVVNDIERS LKTESSPVEA LVLPQKLKDF AKDYEKLVKM HHNVCAMKMR FDKSSDELDP SSSVYEENFK TISKIGNMAK DIINESILKR ESGIHKLQST LDDEKRHLDE EQNMLSEIEF VLSAKDL |
| Entity #616 | Type: protein / Description: DNA-directed RNA polymerase II subunit RPB1 / Formula weight: 12.782 / Num. of mol.: 1 Source: Saccharomyces cerevisiae (strain ATCC 204508 / S288c) References: UniProt: P04050 Sequence: SPEFTCEPTS PSYEPTSPSY EPTSPSYEPT SPSYEPTSPS YEPTSPSYEP TSPSYEPTSP SYEPTSPSYE PTSPSYEPTS PSYEPTSPSY EPTSPSYEPT SPSYEPAAAD YKDDDDK |
-Experimental information
| Beam | Instrument name: CEITEC Rigaku BioSAXS-1000 / City: Brno / 国: Czech Republic  / Type of source: X-ray in house / Wavelength: 0.154 Å / Dist. spec. to detc.: 0.4806 mm / Type of source: X-ray in house / Wavelength: 0.154 Å / Dist. spec. to detc.: 0.4806 mm | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 100K / Pixsize x: 172 mm | |||||||||||||||||||||||||||||||||
| Scan |
| |||||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| |||||||||||||||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller