+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9s3z | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cerebellar GluA1/4 LBD tetramer (focused refinement) | ||||||||||||||||||||||||
 Components Components |
| ||||||||||||||||||||||||
 Keywords Keywords | SIGNALING PROTEIN / AMPA ionotropic glutamate receptor | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationCOPII-mediated vesicle transport / Cargo concentration in the ER / Activation of AMPA receptors / Trafficking of AMPA receptors / Trafficking of GluR2-containing AMPA receptors / Unblocking of NMDA receptors, glutamate binding and activation / axonal spine / cellular response to ammonium ion / dendritic spine membrane / long-term synaptic depression ...COPII-mediated vesicle transport / Cargo concentration in the ER / Activation of AMPA receptors / Trafficking of AMPA receptors / Trafficking of GluR2-containing AMPA receptors / Unblocking of NMDA receptors, glutamate binding and activation / axonal spine / cellular response to ammonium ion / dendritic spine membrane / long-term synaptic depression / perisynaptic space / AMPA glutamate receptor activity / negative regulation of smooth muscle cell apoptotic process / AMPA glutamate receptor complex / long-term memory / synapse assembly / excitatory synapse / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / synaptic transmission, glutamatergic / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / recycling endosome / postsynaptic density membrane / modulation of chemical synaptic transmission / receptor internalization / synaptic vesicle membrane / dendritic spine / neuronal cell body / glutamatergic synapse / cell surface / endoplasmic reticulum / plasma membrane Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  | ||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.55 Å | ||||||||||||||||||||||||
 Authors Authors | Sengupta, N. / Scrutton, A. / Greger, I.H. / Krieger, J.M. | ||||||||||||||||||||||||
| Funding support |  United Kingdom, 2items United Kingdom, 2items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Science / Year: 2025 Journal: Science / Year: 2025Title: Structure and organization of AMPA receptor-TARP complexes in the mammalian cerebellum. Authors: Alexander M Scrutton / Nayanika Sengupta / Josip Ivica / Imogen Stockwell / Sew Peak-Chew / Bishal Singh / Kunimichi Suzuki / Veronica T Chang / Stephen H McLaughlin / James M Krieger / A ...Authors: Alexander M Scrutton / Nayanika Sengupta / Josip Ivica / Imogen Stockwell / Sew Peak-Chew / Bishal Singh / Kunimichi Suzuki / Veronica T Chang / Stephen H McLaughlin / James M Krieger / A Radu Aricescu / Ingo H Greger /   Abstract: AMPA receptors (AMPARs) are multimodal transducers of glutamatergic signals throughout the brain. Their diversity is exemplified in the cerebellum; at afferent synapses, AMPARs mediate high-frequency ...AMPA receptors (AMPARs) are multimodal transducers of glutamatergic signals throughout the brain. Their diversity is exemplified in the cerebellum; at afferent synapses, AMPARs mediate high-frequency excitation, whereas in Bergmann glia (BG) they support calcium transients that modulate synaptic transmission. This spectrum arises from different combinations of core subunits (GluA1-4), auxiliary proteins, and post-transcriptional modifications. Here, using mass-spectrometry, cryo-EM, and electrophysiology, we characterize major cerebellar AMPARs in pig: calcium-impermeable GluA2/A4 heteromers with four TARP subunits, mainly neuronal in origin, and BG-specific calcium-permeable GluA1/A4 heteromers containing two Type-2 TARPs. We also showed that GluA4 receptors consistently exhibit compact N-terminal domains that promote their synaptic delivery. Our study defines the organizational principles of mammalian cerebellar AMPAR complexes and reveals how different receptor subtypes support cell-type specific functions. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9s3z.cif.gz 9s3z.cif.gz | 241.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9s3z.ent.gz pdb9s3z.ent.gz | 164.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  9s3z.json.gz 9s3z.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/s3/9s3z https://data.pdbj.org/pub/pdb/validation_reports/s3/9s3z ftp://data.pdbj.org/pub/pdb/validation_reports/s3/9s3z ftp://data.pdbj.org/pub/pdb/validation_reports/s3/9s3z | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  54556MC  9s3oC  9s3qC  9s41C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
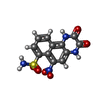
| #1: Protein | Mass: 99762.766 Da / Num. of mol.: 2 / Source method: isolated from a natural source Details: Uniprot A0A286ZS63 (GRIA1 PIG), start 19 after signal peptide cleavage. Source: (natural)  #2: Protein | Mass: 96993.750 Da / Num. of mol.: 2 / Source method: isolated from a natural source Details: Uniprot I3L8N9 (GRIA4 PIG), start 22 after signal peptide cleavage. Source: (natural)  #3: Chemical | ChemComp-E2Q / Has ligand of interest | Y | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Cerebellar GluA1/4 LBD tetramer (focused refinement) / Type: COMPLEX / Entity ID: #1-#2 / Source: NATURAL |
|---|---|
| Source (natural) | Organism:  |
| Buffer solution | pH: 8 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: TFS KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2400 nm / Nominal defocus min: 1200 nm / Cs: 2.7 mm |
| Image recording | Electron dose: 40 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) |
| EM imaging optics | Energyfilter name: GIF Bioquantum |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C2 (2 fold cyclic) | ||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.55 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 329869 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||
| Atomic model building | 3D fitting-ID: 1
| ||||||||||||||||||||||||||||||||||||
| Refinement | Highest resolution: 3.55 Å Stereochemistry target values: REAL-SPACE (WEIGHTED MAP SUM AT ATOM CENTERS) | ||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj