+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cerebellar GluA1/4 LBD tetramer (focused refinement) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AMPA ionotropic glutamate receptor / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationCOPII-mediated vesicle transport / Cargo concentration in the ER / Activation of AMPA receptors / Trafficking of AMPA receptors / Trafficking of GluR2-containing AMPA receptors / Unblocking of NMDA receptors, glutamate binding and activation / axonal spine / cellular response to ammonium ion / dendritic spine membrane / long-term synaptic depression ...COPII-mediated vesicle transport / Cargo concentration in the ER / Activation of AMPA receptors / Trafficking of AMPA receptors / Trafficking of GluR2-containing AMPA receptors / Unblocking of NMDA receptors, glutamate binding and activation / axonal spine / cellular response to ammonium ion / dendritic spine membrane / long-term synaptic depression / perisynaptic space / AMPA glutamate receptor activity / negative regulation of smooth muscle cell apoptotic process / AMPA glutamate receptor complex / long-term memory / synapse assembly / excitatory synapse / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / synaptic transmission, glutamatergic / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / recycling endosome / postsynaptic density membrane / modulation of chemical synaptic transmission / receptor internalization / synaptic vesicle membrane / dendritic spine / neuronal cell body / glutamatergic synapse / cell surface / endoplasmic reticulum / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.55 Å | |||||||||
 Authors Authors | Sengupta N / Scrutton A / Greger IH / Krieger JM | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2025 Journal: Science / Year: 2025Title: Structure and organization of AMPA receptor-TARP complexes in the mammalian cerebellum. Authors: Alexander M Scrutton / Nayanika Sengupta / Josip Ivica / Imogen Stockwell / Sew Peak-Chew / Bishal Singh / Kunimichi Suzuki / Veronica T Chang / Stephen H McLaughlin / James M Krieger / A ...Authors: Alexander M Scrutton / Nayanika Sengupta / Josip Ivica / Imogen Stockwell / Sew Peak-Chew / Bishal Singh / Kunimichi Suzuki / Veronica T Chang / Stephen H McLaughlin / James M Krieger / A Radu Aricescu / Ingo H Greger /   Abstract: AMPA receptors (AMPARs) are multimodal transducers of glutamatergic signals throughout the brain. Their diversity is exemplified in the cerebellum; at afferent synapses, AMPARs mediate high-frequency ...AMPA receptors (AMPARs) are multimodal transducers of glutamatergic signals throughout the brain. Their diversity is exemplified in the cerebellum; at afferent synapses, AMPARs mediate high-frequency excitation, whereas in Bergmann glia (BG) they support calcium transients that modulate synaptic transmission. This spectrum arises from different combinations of core subunits (GluA1-4), auxiliary proteins, and post-transcriptional modifications. Here, using mass-spectrometry, cryo-EM, and electrophysiology, we characterize major cerebellar AMPARs in pig: calcium-impermeable GluA2/A4 heteromers with four TARP subunits, mainly neuronal in origin, and BG-specific calcium-permeable GluA1/A4 heteromers containing two Type-2 TARPs. We also showed that GluA4 receptors consistently exhibit compact N-terminal domains that promote their synaptic delivery. Our study defines the organizational principles of mammalian cerebellar AMPAR complexes and reveals how different receptor subtypes support cell-type specific functions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_54556.map.gz emd_54556.map.gz | 306.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-54556-v30.xml emd-54556-v30.xml emd-54556.xml emd-54556.xml | 20 KB 20 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_54556_fsc.xml emd_54556_fsc.xml | 14.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_54556.png emd_54556.png | 31.2 KB | ||
| Filedesc metadata |  emd-54556.cif.gz emd-54556.cif.gz | 7 KB | ||
| Others |  emd_54556_half_map_1.map.gz emd_54556_half_map_1.map.gz emd_54556_half_map_2.map.gz emd_54556_half_map_2.map.gz | 301.3 MB 301.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-54556 http://ftp.pdbj.org/pub/emdb/structures/EMD-54556 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-54556 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-54556 | HTTPS FTP |
-Related structure data
| Related structure data |  9s3zMC  9s3oC  9s3qC  9s41C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_54556.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_54556.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.826 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_54556_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_54556_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cerebellar GluA1/4 LBD tetramer (focused refinement)
| Entire | Name: Cerebellar GluA1/4 LBD tetramer (focused refinement) |
|---|---|
| Components |
|
-Supramolecule #1: Cerebellar GluA1/4 LBD tetramer (focused refinement)
| Supramolecule | Name: Cerebellar GluA1/4 LBD tetramer (focused refinement) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Glutamate receptor 1
| Macromolecule | Name: Glutamate receptor 1 / type: protein_or_peptide / ID: 1 Details: Uniprot A0A286ZS63 (GRIA1 PIG), start 19 after signal peptide cleavage. Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 99.762766 KDa |
| Sequence | String: ANFPNNIQIG GLFPNQQSQE HAAFRFALSQ LTEPPKLLPQ IDIVNISDSF EMTYRFCSQF SKGVYAIFGF YERRTVNMLT SFCGALHVC FITPSFPVDT SNQFVLQLRP ELQDALISII DHYKWQKFVY IYDADRGLSV LQKVLDTAAE KNWQVTAVNI L TTTEEGYR ...String: ANFPNNIQIG GLFPNQQSQE HAAFRFALSQ LTEPPKLLPQ IDIVNISDSF EMTYRFCSQF SKGVYAIFGF YERRTVNMLT SFCGALHVC FITPSFPVDT SNQFVLQLRP ELQDALISII DHYKWQKFVY IYDADRGLSV LQKVLDTAAE KNWQVTAVNI L TTTEEGYR MLFQDLEKKK ERLVVVDCES ERLNAILGQI IKLEKNGIGY HYILANLGFM DIDLNKFKES GANVTGFQLV NY TDTIPAK IMQQWKTSDA RDHTRVDWKR PKYTSALTYD GVKVMAEAFQ SLRRQRIDIS RRGNAGDCLA NPAVPWGQGI DIQ RALQQV RFEGLTGNVQ FNEKGRRTNY TLHVIEMKHD GIRKIGYWNE DDKFVPAATD AQAGGDNSSV QNRTYIVTTI LEDP YVMLK KNANQFEGND RYEGYCVELA AEIAKHVGYS YRLEIVSDGK YGARDPDTKA WNGMVGELVY GRADVAVAPL TITLV REEV IDFSKPFMSL GISIMIKKPQ KSKPGVFSFL DPLAYEIWMC IVFAYIGVSV VLFLVSRFSP YEWHSEEFEE GRDQTT SDQ SNEFGIFNSL WFSLGAFMQQ GCDISPRSLS GRIVGGVWWF FTLIIISSYT ANLAAFLTVE RMVSPIESAE DLAKQTE IA YGTLEAGSTK EFFRRSKIAV FEKMWTYMKS AEPSVFVRTT EEGMIRVRKS KGKYAYLLES TMNEYIEQRK PCDTMKVG G NLDSKGYGIA TPKGSALRNP VNLAVLKLNE QGLLDKLKNK WWYDKGECGS KDSGSKDKTS ALSLSNVAGV FYILIGGLG LAMLVALIEF CYKSRSESKR MKGFCLIPQQ SINEAIRTST LPRNSGAGAS GGGSGENGRV VSHDFPKSMQ SIPCMSHSTG MPLGATGL UniProtKB: Glutamate receptor |
-Macromolecule #2: Glutamate receptor 4
| Macromolecule | Name: Glutamate receptor 4 / type: protein_or_peptide / ID: 2 Details: Uniprot I3L8N9 (GRIA4 PIG), start 22 after signal peptide cleavage. Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 96.99375 KDa |
| Sequence | String: AFPSSVQIGG LFIRNTDQEY TAFRLAIFLH NTSPNASEAP FNLVPHVDNI ETANSFAVTN AFCSQYSRGV FAIFGLYDKR SVHTLTSFC SALHISLITP SFPTEGESQF VLQLRPSLRG ALLSLLDHYE WNCFVFLYDT DRGYSILQAI MEKAGQNGWH V SAICVENF ...String: AFPSSVQIGG LFIRNTDQEY TAFRLAIFLH NTSPNASEAP FNLVPHVDNI ETANSFAVTN AFCSQYSRGV FAIFGLYDKR SVHTLTSFC SALHISLITP SFPTEGESQF VLQLRPSLRG ALLSLLDHYE WNCFVFLYDT DRGYSILQAI MEKAGQNGWH V SAICVENF NDVSYRQLLE ELDRRQEKKF VIDCEIERLQ NILEQIVSVG KHVKGYHYII ANLGFKDISL ERFIHGGANV TG FQLVDFN TPMVTKLMDR WKKLDQREYP GSETPPKYTS ALTYDGVLVM AETFRNLRRQ KIDISRRGNA GDCLANPAAP WGQ GIDMER TLKQVQIQGL TGNVQFDHYG RRVNYTMDVF ELKSTGPRKV GYWNDMDKLV LIQDVPTLGN DTAAIENRTV VVTT IMESP YVMYKKNHEM FEGNDKYEGY CVDLASEIAK HIGIKYKIAI VPDGKYGARD ADTKIWNGMV GELVYGKAEI AIAPL TITL VREEVIDFSK PFMSLGISIM IKKPQKSKPG VFSFLDPLAY EIWMCIVFAY IGVSVVLFLV SRFSPYEWHT EEPEDG KEG PSDQPPNEFG IFNSLWFSLG AFMQQGCDIS PRSLSGRIVG GVWWFFTLII ISSYTANLAA FLTVERMVSP IESAEDL AK QTEIAYGTLD SGSTKEFFRR SKIAVYEKMW TYMRSAEPSV FTRTTAEGVA RVRKSKGKFA FLLESTMNEY IEQRKPCD T MKVGGNLDSK GYGVATPKGS SLRTPVNLAV LKLSEAGVLD KLKNKWWYDK GECGPKDSGS KDKTSALSLS NVAGVFYIL VGGLGLAMLV ALIEFCYKSR AEAKRMKVAK SAQTFNPTSS QNTQNLATYR EGYNVYGTES IKI UniProtKB: Glutamate receptor |
-Macromolecule #3: 6-nitro-2,3-bis(oxidanylidene)-1,4-dihydrobenzo[f]quinoxaline-7-s...
| Macromolecule | Name: 6-nitro-2,3-bis(oxidanylidene)-1,4-dihydrobenzo[f]quinoxaline-7-sulfonamide type: ligand / ID: 3 / Number of copies: 4 / Formula: E2Q |
|---|---|
| Molecular weight | Theoretical: 336.28 Da |
| Chemical component information | 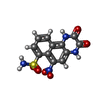 ChemComp-E2Q: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)







































