[English] 日本語
 Yorodumi
Yorodumi- PDB-9rsj: Cryo-EM structure of MATE transporter NorM-VC in complex with dox... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9rsj | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of MATE transporter NorM-VC in complex with doxorubicin | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | TRANSPORT PROTEIN / NabFab / substrate / multidrug transporter | |||||||||
| Function / homology |  Function and homology information Function and homology informationantiporter activity / xenobiotic transmembrane transporter activity / monoatomic ion transport / plasma membrane Similarity search - Function | |||||||||
| Biological species |  synthetic construct (others)  | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Romane, K. / Hsieh, P.Y. / Kowal, J. / Locher, K.P. / van Veen, H.W. | |||||||||
| Funding support |  Switzerland, Switzerland,  United Kingdom, 2items United Kingdom, 2items
| |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2026 Journal: J Mol Biol / Year: 2026Title: Doxorubicin Recognition and Transport by the MATE Multidrug Transporter NorM From Vibrio cholerae. Authors: Pei-Yu Hsieh / Ksenija Romane / Julia Kowal / Kaspar P Locher / Hendrik W van Veen /   Abstract: Multidrug and toxic compound extrusion (MATE) transport proteins contribute to multidrug resistance in human pathogens by extruding various cytotoxic compounds from the cellular interior. Despite ...Multidrug and toxic compound extrusion (MATE) transport proteins contribute to multidrug resistance in human pathogens by extruding various cytotoxic compounds from the cellular interior. Despite their importance across all domains of life, the specificities and mechanisms of substrate transport of these proteins remain poorly understood due to limited structural and functional information. Here, we determined the cryo-electron microscopy structure of NorM from Vibrio cholerae (NorM-VC) in complex with the anthracycline antibiotic doxorubicin, using the NabFab approach. The structure reveals that the doxorubicin-binding pocket is located halfway through the membrane, within the C-lobe of the protein. Functional studies targeting the doxorubicin-interacting residues validated the binding pocket and enabled detailed analysis of the doxorubicin transport reaction. Our findings indicate doxorubicin binding within a multisite binding chamber engaged in a general transport mechanism for a variety of substrates. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9rsj.cif.gz 9rsj.cif.gz | 231 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9rsj.ent.gz pdb9rsj.ent.gz | 179.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  9rsj.json.gz 9rsj.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/rs/9rsj https://data.pdbj.org/pub/pdb/validation_reports/rs/9rsj ftp://data.pdbj.org/pub/pdb/validation_reports/rs/9rsj ftp://data.pdbj.org/pub/pdb/validation_reports/rs/9rsj | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  54220MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Antibody , 4 types, 4 molecules HLNK
| #2: Antibody | Mass: 25684.463 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.) synthetic construct (others) / Production host:  |
|---|---|
| #3: Antibody | Mass: 23258.783 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.) synthetic construct (others) / Production host:  |
| #4: Antibody | Mass: 14356.979 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: Nanobody / Source: (gene. exp.) synthetic construct (others) / Production host:  |
| #5: Antibody | Mass: 13390.644 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: Nanobody / Source: (gene. exp.)   |
-Protein / Non-polymers , 2 types, 2 molecules A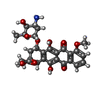

| #1: Protein | Mass: 49892.328 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #6: Chemical | ChemComp-DM2 / |
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: MATE transporter NorM from Vibrio cholerae in complex with NabFab, Nb17_4, anti-Fab nanobody and doxorubicin Type: COMPLEX / Entity ID: #1-#5 / Source: RECOMBINANT |
|---|---|
| Molecular weight | Value: 0.128 MDa / Experimental value: NO |
| Source (natural) | Organism:  |
| Source (recombinant) | Organism:  |
| Buffer solution | pH: 7.4 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE-PROPANE / Humidity: 100 % |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: TFS KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2400 nm / Nominal defocus min: 600 nm |
| Specimen holder | Cryogen: NITROGEN |
| Image recording | Electron dose: 65 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 327809 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refinement | Highest resolution: 3.1 Å / Cross valid method: NONE Stereochemistry target values: REAL-SPACE (WEIGHTED MAP SUM AT ATOM CENTERS) | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj


