[English] 日本語
 Yorodumi
Yorodumi- EMDB-54220: Cryo-EM structure of MATE transporter NorM-VC in complex with dox... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of MATE transporter NorM-VC in complex with doxorubicin | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NabFab / substrate / multidrug transporter / TRANSPORT PROTEIN | |||||||||
| Function / homology | : / : / Multi antimicrobial extrusion protein / MatE / antiporter activity / xenobiotic transmembrane transporter activity / monoatomic ion transport / plasma membrane / Multidrug resistance protein NorM Function and homology information Function and homology information | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Romane K / Hsieh PY / Kowal J / Locher KP / van Veen HW | |||||||||
| Funding support |  Switzerland, Switzerland,  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2026 Journal: J Mol Biol / Year: 2026Title: Doxorubicin Recognition and Transport by the MATE Multidrug Transporter NorM From Vibrio cholerae. Authors: Pei-Yu Hsieh / Ksenija Romane / Julia Kowal / Kaspar P Locher / Hendrik W van Veen /   Abstract: Multidrug and toxic compound extrusion (MATE) transport proteins contribute to multidrug resistance in human pathogens by extruding various cytotoxic compounds from the cellular interior. Despite ...Multidrug and toxic compound extrusion (MATE) transport proteins contribute to multidrug resistance in human pathogens by extruding various cytotoxic compounds from the cellular interior. Despite their importance across all domains of life, the specificities and mechanisms of substrate transport of these proteins remain poorly understood due to limited structural and functional information. Here, we determined the cryo-electron microscopy structure of NorM from Vibrio cholerae (NorM-VC) in complex with the anthracycline antibiotic doxorubicin, using the NabFab approach. The structure reveals that the doxorubicin-binding pocket is located halfway through the membrane, within the C-lobe of the protein. Functional studies targeting the doxorubicin-interacting residues validated the binding pocket and enabled detailed analysis of the doxorubicin transport reaction. Our findings indicate doxorubicin binding within a multisite binding chamber engaged in a general transport mechanism for a variety of substrates. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_54220.map.gz emd_54220.map.gz | 307.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-54220-v30.xml emd-54220-v30.xml emd-54220.xml emd-54220.xml | 21.1 KB 21.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_54220_fsc.xml emd_54220_fsc.xml | 14.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_54220.png emd_54220.png | 95 KB | ||
| Filedesc metadata |  emd-54220.cif.gz emd-54220.cif.gz | 6.6 KB | ||
| Others |  emd_54220_half_map_1.map.gz emd_54220_half_map_1.map.gz emd_54220_half_map_2.map.gz emd_54220_half_map_2.map.gz | 302.8 MB 302.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-54220 http://ftp.pdbj.org/pub/emdb/structures/EMD-54220 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-54220 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-54220 | HTTPS FTP |
-Related structure data
| Related structure data |  9rsjMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_54220.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_54220.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.67 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_54220_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_54220_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : MATE transporter NorM from Vibrio cholerae in complex with NabFab...
| Entire | Name: MATE transporter NorM from Vibrio cholerae in complex with NabFab, Nb17_4, anti-Fab nanobody and doxorubicin |
|---|---|
| Components |
|
-Supramolecule #1: MATE transporter NorM from Vibrio cholerae in complex with NabFab...
| Supramolecule | Name: MATE transporter NorM from Vibrio cholerae in complex with NabFab, Nb17_4, anti-Fab nanobody and doxorubicin type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 128 KDa |
-Macromolecule #1: Multidrug resistance protein NorM
| Macromolecule | Name: Multidrug resistance protein NorM / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49.892328 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MENSVHRYKK EASNLIKLAT PVLIASVAQT GMGFVDTIMA GGVSAIDMAA VSIAASIWLP SILFGVGLLM ALVPVVAQLN GAGRQHKIP FEVHQGLILA LLVSVPIIAV LFQTQFIIRF MDVEEAMATK TVGYMHAVIF AVPAYLLFQA LRSFTDGMSL T KPAMVIGF ...String: MENSVHRYKK EASNLIKLAT PVLIASVAQT GMGFVDTIMA GGVSAIDMAA VSIAASIWLP SILFGVGLLM ALVPVVAQLN GAGRQHKIP FEVHQGLILA LLVSVPIIAV LFQTQFIIRF MDVEEAMATK TVGYMHAVIF AVPAYLLFQA LRSFTDGMSL T KPAMVIGF IGLLLNIPLN WIFVYGKFGA PELGGVGCGV ATAIVYWIML LLLLFYIVTS KRLAHVKVFE TFHKPQPKEL IR LFRLGFP VAAALFFEVT LFAVVALLVA PLGSTVVAAH QVALNFSSLV FMFPMSIGAA VSIRVGHKLG EQDTKGAAIA ANV GLMTGL ATACITALLT VLFREQIALL YTENQVVVAL AMQLLLFAAI YQCMDAVQVV AAGSLRGYKD MTAIFHRTFI SYWV LGLPT GYILGMTNWL TEQPLGAKGF WLGFIIGLSA AALMLGQRLY WLQKQSDDVQ LHLAAK UniProtKB: Multidrug resistance protein NorM |
-Macromolecule #2: NabFab HC
| Macromolecule | Name: NabFab HC / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 25.684463 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NFSYYSIHWV RQAPGKGLEW VAYISSSSSY TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARGYQYWQYH ASWYWNGGLD YWGQGTLVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL V KDYFPEPV ...String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NFSYYSIHWV RQAPGKGLEW VAYISSSSSY TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARGYQYWQYH ASWYWNGGLD YWGQGTLVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL V KDYFPEPV TVSWNSGALT SGVHTFPAVL QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKK VEPKSCDKTH T |
-Macromolecule #3: NabFab LC
| Macromolecule | Name: NabFab LC / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 23.258783 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQSSSSLITF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQSSSSLITF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #4: NorM-Nb17_4
| Macromolecule | Name: NorM-Nb17_4 / type: protein_or_peptide / ID: 4 / Details: Nanobody / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 14.356979 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QRQLVESGGG LVQPGGSLRL SCAASGIIFK INDMGWFRQA PGKEREGVAG ITSGGRTNYA DSVKGRFIIS RDNVKNTVYL QMNSLEPED TAVYYCKSDG LISYAASQLS TYWGKGTPVT VSSHHHHHHE PEA |
-Macromolecule #5: Anti-Fab nanobody
| Macromolecule | Name: Anti-Fab nanobody / type: protein_or_peptide / ID: 5 / Details: Nanobody / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.390644 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSQVQLQESG GGLVQPGGSL RLSCAASGRT ISRYAMSWFR QAPGKEREFV AVARRSGDGA FYADSVQGRF TVSRDDAKNT VYLQMNSLK PEDTAVYYCA IDSDTFYSGS YDYWGQGTQV TVSS |
-Macromolecule #6: DOXORUBICIN
| Macromolecule | Name: DOXORUBICIN / type: ligand / ID: 6 / Number of copies: 1 / Formula: DM2 |
|---|---|
| Molecular weight | Theoretical: 543.519 Da |
| Chemical component information | 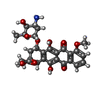 ChemComp-DM2: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 65.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.6 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Software | Name:  Coot Coot |
|---|---|
| Output model |  PDB-9rsj: |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































