[English] 日本語
 Yorodumi
Yorodumi- PDB-9o8u: (1-methylalkyl)succinate synthase alpha-beta-gamma-delta complex ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9o8u | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | (1-methylalkyl)succinate synthase alpha-beta-gamma-delta complex with bound fumarate | ||||||||||||||||||||||||||||||
 Components Components |
| ||||||||||||||||||||||||||||||
 Keywords Keywords | LYASE / glycyl radical enzyme / (1-methylalkyl)succinate synthase / X-succinate synthase / alkylsuccinate synthase / Hydrocarbon degradation / fumarate addition | ||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology information | ||||||||||||||||||||||||||||||
| Biological species |  Azoarcus sp. HxN1 (bacteria) Azoarcus sp. HxN1 (bacteria) | ||||||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.8 Å | ||||||||||||||||||||||||||||||
 Authors Authors | Andorfer, M.C. / Drennan, C.L. | ||||||||||||||||||||||||||||||
| Funding support |  United States, 3items United States, 3items
| ||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2025 Journal: Proc Natl Acad Sci U S A / Year: 2025Title: Structural basis for anaerobic alkane activation by a multisubunit glycyl radical enzyme. Authors: Mary C Andorfer / Talya S Levitz / Jian Liu / Ankush Chakraborty / Devin T King-Roberts / Delight Nweneka / Christa N Imrich / Catherine L Drennan /  Abstract: X-succinate synthases (XSSs) are glycyl radical enzymes (GREs) that catalyze the addition of hydrocarbons to fumarate via radical chemistry, thereby activating them for microbial metabolism. To date, ...X-succinate synthases (XSSs) are glycyl radical enzymes (GREs) that catalyze the addition of hydrocarbons to fumarate via radical chemistry, thereby activating them for microbial metabolism. To date, the only structurally characterized XSS is benzylsuccinate synthase (BSS), which functionalizes toluene. A distinct subclass of XSSs acts on saturated hydrocarbons, which possess much stronger C(sp)-H bonds than toluene, suggesting mechanistic and structural differences from BSS. Here, we use cryogenic electron microscopy to determine the structure of one such enzyme, (1-methylalkyl)succinate synthase (MASS) from strain HxN1, which functionalizes -alkanes (C6-C8). The structure reveals an asymmetric dimer in which both sides contain a catalytic α-subunit and accessory γ-subunit. One α-subunit also binds two additional subunits, β and δ. The β-subunit binds a [4Fe-4S] cluster and adopts a fold similar to BSSβ. The β-subunit appears to regulate the flexibility of the α-subunit to enable opening of the active site, affording the binding of -alkane substrates. The δ-subunit, which lacks homology to known GRE subunits, adopts a rubredoxin-like fold that binds a single Fe ion, an architecture not previously reported for GREs. MASSδ occupies the same region of the α-subunit as the activating enzyme (AE) and may regulate the conformational changes required for glycyl radical installation. Structural comparisons between MASS and BSS reveal differences in how fumarate is bound and show amino acid substitutions that could account for the binding of alkanes versus toluene. Together, this structure offers insight into anaerobic alkane activation via fumarate addition. | ||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9o8u.cif.gz 9o8u.cif.gz | 323 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9o8u.ent.gz pdb9o8u.ent.gz | 256.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  9o8u.json.gz 9o8u.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  9o8u_validation.pdf.gz 9o8u_validation.pdf.gz | 1.2 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  9o8u_full_validation.pdf.gz 9o8u_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  9o8u_validation.xml.gz 9o8u_validation.xml.gz | 58.3 KB | Display | |
| Data in CIF |  9o8u_validation.cif.gz 9o8u_validation.cif.gz | 89.8 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/o8/9o8u https://data.pdbj.org/pub/pdb/validation_reports/o8/9o8u ftp://data.pdbj.org/pub/pdb/validation_reports/o8/9o8u ftp://data.pdbj.org/pub/pdb/validation_reports/o8/9o8u | HTTPS FTP |
-Related structure data
| Related structure data |  70238MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-1-methyl alkyl succinate synthase subunit ... , 3 types, 5 molecules AECFD
| #1: Protein | Mass: 97053.289 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Azoarcus sp. HxN1 (bacteria) / Gene: masD / Production host: Azoarcus sp. HxN1 (bacteria) / Gene: masD / Production host:  #3: Protein | Mass: 6894.553 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Azoarcus sp. HxN1 (bacteria) / Gene: masC / Production host: Azoarcus sp. HxN1 (bacteria) / Gene: masC / Production host:  #4: Protein | | Mass: 8033.105 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Azoarcus sp. HxN1 (bacteria) / Gene: masE / Production host: Azoarcus sp. HxN1 (bacteria) / Gene: masE / Production host:  |
|---|
-Protein , 1 types, 1 molecules B
| #2: Protein | Mass: 13460.091 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Azoarcus sp. HxN1 (bacteria) / Gene: masB / Production host: Azoarcus sp. HxN1 (bacteria) / Gene: masB / Production host:  |
|---|
-Non-polymers , 4 types, 6 molecules 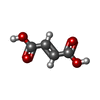






| #5: Chemical | ChemComp-FUM / | ||
|---|---|---|---|
| #6: Chemical | ChemComp-DTT / | ||
| #7: Chemical | | #8: Chemical | ChemComp-FE / | |
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | N |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Heterodimer complex of MASS alpha with beta, gamma, delta subunits Type: COMPLEX Details: one alpha subunits binds beta, gamma, delta subunits, and the other alpha subunit bind only gamma subunits. Entity ID: #1-#4 / Source: RECOMBINANT | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Value: 0.229 MDa / Experimental value: NO | ||||||||||||||||||||
| Source (natural) | Organism:  Azoarcus sp. HxN1 (bacteria) Azoarcus sp. HxN1 (bacteria) | ||||||||||||||||||||
| Source (recombinant) | Organism:  | ||||||||||||||||||||
| Buffer solution | pH: 8 Details: 50 mM HEPES pH 8.0, 300 mM NaCl, 1 mM Fumarate, 1 mM DTT | ||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||
| Specimen | Conc.: 0.8 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||||||||||
| Specimen support | Grid material: COPPER / Grid mesh size: 300 divisions/in. / Grid type: C-flat-1.2/1.3 | ||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: TFS KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: SPOT SCAN FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: SPOT SCAN |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2500 nm / Nominal defocus min: 1000 nm |
| Image recording | Electron dose: 1.42 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) |
- Processing
Processing
| EM software |
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: NONE | ||||||||||||||||
| 3D reconstruction | Resolution: 2.8 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 356678 / Symmetry type: POINT | ||||||||||||||||
| Atomic model building | Protocol: AB INITIO MODEL | ||||||||||||||||
| Atomic model building | Source name: AlphaFold / Type: in silico model |
 Movie
Movie Controller
Controller


 PDBj
PDBj







