[English] 日本語
 Yorodumi
Yorodumi- EMDB-70238: (1-methylalkyl)succinate synthase alpha-beta-gamma-delta complex ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | (1-methylalkyl)succinate synthase alpha-beta-gamma-delta complex with bound fumarate | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | glycyl radical enzyme / (1-methylalkyl)succinate synthase / X-succinate synthase / alkylsuccinate synthase / Hydrocarbon degradation / fumarate addition / LYASE | ||||||||||||
| Function / homology |  Function and homology information Function and homology information | ||||||||||||
| Biological species |  Azoarcus sp. HxN1 (bacteria) Azoarcus sp. HxN1 (bacteria) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | ||||||||||||
 Authors Authors | Andorfer MC / Drennan CL | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2025 Journal: Proc Natl Acad Sci U S A / Year: 2025Title: Structural basis for anaerobic alkane activation by a multisubunit glycyl radical enzyme. Authors: Mary C Andorfer / Talya S Levitz / Jian Liu / Ankush Chakraborty / Devin T King-Roberts / Delight Nweneka / Christa N Imrich / Catherine L Drennan /  Abstract: X-succinate synthases (XSSs) are glycyl radical enzymes (GREs) that catalyze the addition of hydrocarbons to fumarate via radical chemistry, thereby activating them for microbial metabolism. To date, ...X-succinate synthases (XSSs) are glycyl radical enzymes (GREs) that catalyze the addition of hydrocarbons to fumarate via radical chemistry, thereby activating them for microbial metabolism. To date, the only structurally characterized XSS is benzylsuccinate synthase (BSS), which functionalizes toluene. A distinct subclass of XSSs acts on saturated hydrocarbons, which possess much stronger C(sp)-H bonds than toluene, suggesting mechanistic and structural differences from BSS. Here, we use cryogenic electron microscopy to determine the structure of one such enzyme, (1-methylalkyl)succinate synthase (MASS) from strain HxN1, which functionalizes -alkanes (C6-C8). The structure reveals an asymmetric dimer in which both sides contain a catalytic α-subunit and accessory γ-subunit. One α-subunit also binds two additional subunits, β and δ. The β-subunit binds a [4Fe-4S] cluster and adopts a fold similar to BSSβ. The β-subunit appears to regulate the flexibility of the α-subunit to enable opening of the active site, affording the binding of -alkane substrates. The δ-subunit, which lacks homology to known GRE subunits, adopts a rubredoxin-like fold that binds a single Fe ion, an architecture not previously reported for GREs. MASSδ occupies the same region of the α-subunit as the activating enzyme (AE) and may regulate the conformational changes required for glycyl radical installation. Structural comparisons between MASS and BSS reveal differences in how fumarate is bound and show amino acid substitutions that could account for the binding of alkanes versus toluene. Together, this structure offers insight into anaerobic alkane activation via fumarate addition. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_70238.map.gz emd_70238.map.gz | 5.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-70238-v30.xml emd-70238-v30.xml emd-70238.xml emd-70238.xml | 27.8 KB 27.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_70238_fsc.xml emd_70238_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_70238.png emd_70238.png | 152.2 KB | ||
| Masks |  emd_70238_msk_1.map emd_70238_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-70238.cif.gz emd-70238.cif.gz | 7.5 KB | ||
| Others |  emd_70238_additional_1.map.gz emd_70238_additional_1.map.gz emd_70238_half_map_1.map.gz emd_70238_half_map_1.map.gz emd_70238_half_map_2.map.gz emd_70238_half_map_2.map.gz | 49.7 MB 49.8 MB 49.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-70238 http://ftp.pdbj.org/pub/emdb/structures/EMD-70238 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-70238 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-70238 | HTTPS FTP |
-Related structure data
| Related structure data |  9o8uMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_70238.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_70238.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.17 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_70238_msk_1.map emd_70238_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_70238_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_70238_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_70238_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Heterodimer complex of MASS alpha with beta, gamma, delta subunits
| Entire | Name: Heterodimer complex of MASS alpha with beta, gamma, delta subunits |
|---|---|
| Components |
|
-Supramolecule #1: Heterodimer complex of MASS alpha with beta, gamma, delta subunits
| Supramolecule | Name: Heterodimer complex of MASS alpha with beta, gamma, delta subunits type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: one alpha subunits binds beta, gamma, delta subunits, and the other alpha subunit bind only gamma subunits. |
|---|---|
| Source (natural) | Organism:  Azoarcus sp. HxN1 (bacteria) Azoarcus sp. HxN1 (bacteria) |
| Molecular weight | Theoretical: 229 KDa |
-Macromolecule #1: 1-methyl alkyl succinate synthase subunit MasD
| Macromolecule | Name: 1-methyl alkyl succinate synthase subunit MasD / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Azoarcus sp. HxN1 (bacteria) Azoarcus sp. HxN1 (bacteria) |
| Molecular weight | Theoretical: 97.053289 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SQMTATSTLS KTDLKNCETV EELRENQQWW WLAERERSAR LDYLRKATWK KGALGGNYFD GIRLDLEYPT LFTEAWKKY PNDPSMLRRA KATAYVLDNI SIFITDSAQL VGYVGSAPHT IAWRVDGAST VNSEVYNEPG IHAEPEAESL K KVAEINSY ...String: MGSSHHHHHH SQMTATSTLS KTDLKNCETV EELRENQQWW WLAERERSAR LDYLRKATWK KGALGGNYFD GIRLDLEYPT LFTEAWKKY PNDPSMLRRA KATAYVLDNI SIFITDSAQL VGYVGSAPHT IAWRVDGAST VNSEVYNEPG IHAEPEAESL K KVAEINSY WNGQTAVDKV GRLIDPEDAV KFFSGAIGWG TPSSAFGYSG KNFEYFMKGD RAFSQIIAEI DEKIDEAEEA TI GTPSPHI LPLYDKLNNW HAMKLVLEAA IRFAGRYARL ARVMAAKETD EQRKKELLRV AETCERVPAN PPRNLQESLQ YEH FVQVLA RYEAHEGAWP SRPDYYHGPL YAKDVEVEKN ITESEAIDLV GEYMIRCSEY GSFSPRYMRE GLQGVTGTFV WTLG GVNQD GTDACNGMTI ALLKAARLVR VANPTFGFRW HPKVSNEVLR ECFECIRQGL GYPTLRNDPV LIQNTMHWYG HPLEE ARTW VHMACMSPNP TTKHGTSPFR MASATMNSAK TIEYVLHNGY DRVVNMQMGP KTGDAREIKD FEDLFERWTV QLKWLM NLL VRTVNLGRFK DPEFFGRPFL SAITERAVEH GIDAVSPEGE RGNAWVTAFT WIENVDSMAA IKKLVFDDKK YTMSQLI DA LEAEWDGYEQ MRLDFVKNGP KWGNDDDYVD DIMLRCLSVA AEHSRNIQCT SGNCWPILPE NVSGNIHYAN IVGALPNG R RRGDALYDGG VSPGPGLDKA GPTAVLKSVG KIDHVNQGRS FLLNQRLSPT QLAGDKGFQL WNSYVRTWAE LGIDHIQFN VISDKVLRAA QNDPEGYQEV IVRVAGYSAH FIDISRKTQD NIIQRTVQGL GENLYFQG UniProtKB: 1-methyl alkyl succinate synthase subunit MasD |
-Macromolecule #2: MasB protein
| Macromolecule | Name: MasB protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Azoarcus sp. HxN1 (bacteria) Azoarcus sp. HxN1 (bacteria) |
| Molecular weight | Theoretical: 13.460091 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSRRDEWKKL QEEMTRDGGE IKSLETVPEQ ACGICLNFTD NAYGSDGRGS CNVLKAGSNI SLPDVIITRS GENGYITFFN SDAKYCPNF ERMKLIDTDG HECADPISRR VQRQLSSIKK S UniProtKB: MasB protein |
-Macromolecule #3: 1-methyl alkyl succinate synthase subunit MasC
| Macromolecule | Name: 1-methyl alkyl succinate synthase subunit MasC / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Azoarcus sp. HxN1 (bacteria) Azoarcus sp. HxN1 (bacteria) |
| Molecular weight | Theoretical: 6.894553 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: STCKECRNYF PINEEASRGD CVRRISDERQ SYYTARPTTE AAKCEGCSDY LENTRTAKAH UniProtKB: 1-methyl alkyl succinate synthase subunit MasC |
-Macromolecule #4: 1-methyl alkyl succinate synthase subunit Mas E
| Macromolecule | Name: 1-methyl alkyl succinate synthase subunit Mas E / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Azoarcus sp. HxN1 (bacteria) Azoarcus sp. HxN1 (bacteria) |
| Molecular weight | Theoretical: 8.033105 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKCTECGHEA EVMKFRYHYN PRIDASLSLR QCPECQAVVT VDELKREVLG RMHNGDDPWG KSAGIENLAE G UniProtKB: 1-methyl alkyl succinate synthase subunit Mas E |
-Macromolecule #5: FUMARIC ACID
| Macromolecule | Name: FUMARIC ACID / type: ligand / ID: 5 / Number of copies: 1 / Formula: FUM |
|---|---|
| Molecular weight | Theoretical: 116.072 Da |
| Chemical component information | 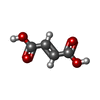 ChemComp-FUM: |
-Macromolecule #6: 2,3-DIHYDROXY-1,4-DITHIOBUTANE
| Macromolecule | Name: 2,3-DIHYDROXY-1,4-DITHIOBUTANE / type: ligand / ID: 6 / Number of copies: 1 / Formula: DTT |
|---|---|
| Molecular weight | Theoretical: 154.251 Da |
| Chemical component information |  ChemComp-DTT: |
-Macromolecule #7: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 7 / Number of copies: 3 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Macromolecule #8: FE (III) ION
| Macromolecule | Name: FE (III) ION / type: ligand / ID: 8 / Number of copies: 1 / Formula: FE |
|---|---|
| Molecular weight | Theoretical: 55.845 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: 50 mM HEPES pH 8.0, 300 mM NaCl, 1 mM Fumarate, 1 mM DTT | ||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. | ||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 1.42 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Refinement | Protocol: AB INITIO MODEL |
| Output model |  PDB-9o8u: |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















































