+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9jm0 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | retron Ec86-effector fiber | |||||||||||||||||||||
 Components Components |
| |||||||||||||||||||||
 Keywords Keywords | RNA BINDING PROTEIN/DNA/RNA / Reverse transcriptase / RNA BINDING PROTEIN-DNA-RNA complex | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationRNA-directed DNA polymerase / RNA-directed DNA polymerase activity / defense response to virus / RNA binding / metal ion binding Similarity search - Function | |||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||||||||||||||
 Authors Authors | Wang, Y.J. / Guan, Z.Y. / Wang, C. / Zou, T.T. | |||||||||||||||||||||
| Funding support |  China, 1items China, 1items
| |||||||||||||||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2024 Journal: Cell Rep / Year: 2024Title: DNA methylation activates retron Ec86 filaments for antiphage defense. Authors: Yanjing Wang / Chen Wang / Zeyuan Guan / Jie Cao / Jia Xu / Shuangshuang Wang / Yongqing Cui / Qiang Wang / Yibei Chen / Yongqi Yin / Delin Zhang / Hongbo Liu / Ming Sun / Shuangxia Jin / ...Authors: Yanjing Wang / Chen Wang / Zeyuan Guan / Jie Cao / Jia Xu / Shuangshuang Wang / Yongqing Cui / Qiang Wang / Yibei Chen / Yongqi Yin / Delin Zhang / Hongbo Liu / Ming Sun / Shuangxia Jin / Pan Tao / Tingting Zou /  Abstract: Retrons are a class of multigene antiphage defense systems typically consisting of a retron reverse transcriptase, a non-coding RNA, and a cognate effector. Although triggers for several retron ...Retrons are a class of multigene antiphage defense systems typically consisting of a retron reverse transcriptase, a non-coding RNA, and a cognate effector. Although triggers for several retron systems have been discovered recently, the complete mechanism by which these systems detect invading phages and mediate defense remains unclear. Here, we focus on the retron Ec86 defense system, elucidating its modes of activation and mechanisms of action. We identified a phage-encoded DNA cytosine methyltransferase (Dcm) as a trigger of the Ec86 system and demonstrated that Ec86 is activated upon multicopy single-stranded DNA (msDNA) methylation. We further elucidated the structure of a tripartite retron Ec86-effector filament assembly that is primed for activation by Dcm and capable of hydrolyzing nicotinamide adenine dinucleotide (NAD). These findings provide insights into the retron Ec86 defense mechanism and underscore an emerging theme of antiphage defense through supramolecular complex assemblies. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9jm0.cif.gz 9jm0.cif.gz | 703.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9jm0.ent.gz pdb9jm0.ent.gz | 561.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  9jm0.json.gz 9jm0.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/jm/9jm0 https://data.pdbj.org/pub/pdb/validation_reports/jm/9jm0 ftp://data.pdbj.org/pub/pdb/validation_reports/jm/9jm0 ftp://data.pdbj.org/pub/pdb/validation_reports/jm/9jm0 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  61595MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 2 types, 8 molecules ABKPCJLT
| #1: Protein | Mass: 37832.070 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   #2: Protein | Mass: 35538.242 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|
-DNA chain , 1 types, 4 molecules DGMQ
| #3: DNA chain | Mass: 26320.875 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|
-RNA chain , 2 types, 8 molecules EHNRFIOS
| #4: RNA chain | Mass: 26199.410 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Details: msrRNA / Source: (gene. exp.)   #5: RNA chain | Mass: 4541.771 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|
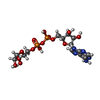
-Non-polymers , 2 types, 2 molecules 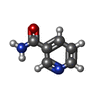

| #6: Chemical | ChemComp-NCA / |
|---|---|
| #7: Chemical | ChemComp-AR6 / [( |
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: retron Ec86-effector fiber / Type: COMPLEX / Entity ID: #1-#5 / Source: MULTIPLE SOURCES |
|---|---|
| Source (natural) | Organism:  |
| Source (recombinant) | Organism:  |
| Buffer solution | pH: 8 |
| Specimen | Conc.: 1 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TECNAI SPIRIT |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2200 nm / Nominal defocus min: 1200 nm |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) |
- Processing
Processing
| EM software | Name: PHENIX / Category: model refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: NONE | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 2.7 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 270375 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj


































































