+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | retron Ec86-effector fiber | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Reverse transcriptase / RNA BINDING PROTEIN/DNA/RNA / RNA BINDING PROTEIN-DNA-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationRNA-directed DNA polymerase / RNA-directed DNA polymerase activity / defense response to virus / RNA binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Wang YJ / Guan ZY / Wang C / Zou TT | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2024 Journal: Cell Rep / Year: 2024Title: DNA methylation activates retron Ec86 filaments for antiphage defense. Authors: Yanjing Wang / Chen Wang / Zeyuan Guan / Jie Cao / Jia Xu / Shuangshuang Wang / Yongqing Cui / Qiang Wang / Yibei Chen / Yongqi Yin / Delin Zhang / Hongbo Liu / Ming Sun / Shuangxia Jin / ...Authors: Yanjing Wang / Chen Wang / Zeyuan Guan / Jie Cao / Jia Xu / Shuangshuang Wang / Yongqing Cui / Qiang Wang / Yibei Chen / Yongqi Yin / Delin Zhang / Hongbo Liu / Ming Sun / Shuangxia Jin / Pan Tao / Tingting Zou /  Abstract: Retrons are a class of multigene antiphage defense systems typically consisting of a retron reverse transcriptase, a non-coding RNA, and a cognate effector. Although triggers for several retron ...Retrons are a class of multigene antiphage defense systems typically consisting of a retron reverse transcriptase, a non-coding RNA, and a cognate effector. Although triggers for several retron systems have been discovered recently, the complete mechanism by which these systems detect invading phages and mediate defense remains unclear. Here, we focus on the retron Ec86 defense system, elucidating its modes of activation and mechanisms of action. We identified a phage-encoded DNA cytosine methyltransferase (Dcm) as a trigger of the Ec86 system and demonstrated that Ec86 is activated upon multicopy single-stranded DNA (msDNA) methylation. We further elucidated the structure of a tripartite retron Ec86-effector filament assembly that is primed for activation by Dcm and capable of hydrolyzing nicotinamide adenine dinucleotide (NAD). These findings provide insights into the retron Ec86 defense mechanism and underscore an emerging theme of antiphage defense through supramolecular complex assemblies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_61595.map.gz emd_61595.map.gz | 79.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-61595-v30.xml emd-61595-v30.xml emd-61595.xml emd-61595.xml | 21.6 KB 21.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_61595.png emd_61595.png | 44.7 KB | ||
| Filedesc metadata |  emd-61595.cif.gz emd-61595.cif.gz | 6.8 KB | ||
| Others |  emd_61595_half_map_1.map.gz emd_61595_half_map_1.map.gz emd_61595_half_map_2.map.gz emd_61595_half_map_2.map.gz | 77.7 MB 77.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-61595 http://ftp.pdbj.org/pub/emdb/structures/EMD-61595 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61595 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61595 | HTTPS FTP |
-Related structure data
| Related structure data |  9jm0MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_61595.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_61595.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_61595_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_61595_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : retron Ec86-effector fiber
| Entire | Name: retron Ec86-effector fiber |
|---|---|
| Components |
|
-Supramolecule #1: retron Ec86-effector fiber
| Supramolecule | Name: retron Ec86-effector fiber / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Retron Ec86 reverse transcriptase
| Macromolecule | Name: Retron Ec86 reverse transcriptase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: RNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.83207 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKSAEYLNTF RLRNLGLPVM NNLHDMSKAT RISVETLRLL IYTADFRYRI YTVEKKGPEK RMRTIYQPSR ELKALQGWVL RNILDKLSS SPFSIGFEKH QSILNNATPH IGANFILNID LEDFFPSLTA NKVFGVFHSL GYNRLISSVL TKICCYKNLL P QGAPSSPK ...String: MKSAEYLNTF RLRNLGLPVM NNLHDMSKAT RISVETLRLL IYTADFRYRI YTVEKKGPEK RMRTIYQPSR ELKALQGWVL RNILDKLSS SPFSIGFEKH QSILNNATPH IGANFILNID LEDFFPSLTA NKVFGVFHSL GYNRLISSVL TKICCYKNLL P QGAPSSPK LANLICSKLD YRIQGYAGSR GLIYTRYADD LTLSAQSMKK VVKARDFLFS IIPSEGLVIN SKKTCISGPR SQ RKVTGLV ISQEKVGIGR EKYKEIRAKI HHIFCGKSSE IEHVRGWLSF ILSVDSKSHR RLITYISKLE KKYGKNPLNK AKT LEHHHH HHHH UniProtKB: Retron Ec86 reverse transcriptase |
-Macromolecule #2: Retron Ec86 putative ribosyltransferase/DNA-binding protein
| Macromolecule | Name: Retron Ec86 putative ribosyltransferase/DNA-binding protein type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 35.538242 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNKKFTDEQQ QQLIGHLTKK GFYRGANIKI TIFLCGGDVA NHQSWRHQLS QFLAKFSDVD IFYPEDLFDD LLAGQGQHSL LSLENILAE AVDVIILFPE SPGSFTELGA FSNNENLRRK LICIQDAKFK SKRSFINYGP VRLLRKFNSK SVLRCSSNEL K EMCDSSID ...String: MNKKFTDEQQ QQLIGHLTKK GFYRGANIKI TIFLCGGDVA NHQSWRHQLS QFLAKFSDVD IFYPEDLFDD LLAGQGQHSL LSLENILAE AVDVIILFPE SPGSFTELGA FSNNENLRRK LICIQDAKFK SKRSFINYGP VRLLRKFNSK SVLRCSSNEL K EMCDSSID VARKLRLYKK LMASIKKVRK ENKVSKDIGN ILYAERFLLP CIYLLDSVNY RTLCELAFKA IKQDDVLSKI IV RSVVSRL INERKILQMT DGYQVTALGA SYVRSVFDRK TLDRLRLEIM NFENRRKSTF NYDKIPYAHP UniProtKB: Retron Ec86 putative ribosyltransferase/DNA-binding protein |
-Macromolecule #3: DNA (85-MER)
| Macromolecule | Name: DNA (85-MER) / type: dna / ID: 3 / Number of copies: 4 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.320875 KDa |
| Sequence | String: (DG)(DT)(DC)(DA)(DG)(DA)(DA)(DA)(DA)(DA) (DA)(DC)(DG)(DG)(DG)(DT)(DT)(DT)(DC)(DC) (DT)(DG)(DG)(DT)(DT)(DG)(DG)(DC)(DT) (DC)(DG)(DG)(DA)(DG)(DA)(DG)(DC)(DA)(DT) (DC) (DA)(DG)(DG)(DC)(DG)(DA) ...String: (DG)(DT)(DC)(DA)(DG)(DA)(DA)(DA)(DA)(DA) (DA)(DC)(DG)(DG)(DG)(DT)(DT)(DT)(DC)(DC) (DT)(DG)(DG)(DT)(DT)(DG)(DG)(DC)(DT) (DC)(DG)(DG)(DA)(DG)(DA)(DG)(DC)(DA)(DT) (DC) (DA)(DG)(DG)(DC)(DG)(DA)(DT)(DG) (DC)(DT)(DC)(DT)(DC)(DC)(DG)(DT)(DT)(DC) (DC)(DA) (DA)(DC)(DA)(DA)(DG)(DG)(DA) (DA)(DA)(DA)(DC)(DA)(DG)(DA)(DC)(DA)(DG) (DT)(DA)(DA) (DC)(DT)(DC)(DA)(DG) GENBANK: GENBANK: M24408.1 |
-Macromolecule #4: RNA (82-MER)
| Macromolecule | Name: RNA (82-MER) / type: rna / ID: 4 / Details: msrRNA / Number of copies: 4 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.19941 KDa |
| Sequence | String: AUGCGCACCC UUAGCGAGAG GUUUAUCAUU AAGGUCAACC UCUGGAUGUU GUUUCGGCAU CCUGCAUUGA AUCUGAGUUA CU GENBANK: GENBANK: M24408.1 |
-Macromolecule #5: RNA (5'-R(P*CP*GP*UP*AP*AP*GP*GP*GP*UP*GP*CP*GP*CP*A)-3')
| Macromolecule | Name: RNA (5'-R(P*CP*GP*UP*AP*AP*GP*GP*GP*UP*GP*CP*GP*CP*A)-3') type: rna / ID: 5 / Number of copies: 4 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 4.541771 KDa |
| Sequence | String: CGUAAGGGUG CGCA |
-Macromolecule #6: NICOTINAMIDE
| Macromolecule | Name: NICOTINAMIDE / type: ligand / ID: 6 / Number of copies: 1 / Formula: NCA |
|---|---|
| Molecular weight | Theoretical: 122.125 Da |
| Chemical component information | 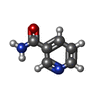 ChemComp-NCA: |
-Macromolecule #7: [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]ME...
| Macromolecule | Name: [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL[HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE type: ligand / ID: 7 / Number of copies: 1 / Formula: AR6 |
|---|---|
| Molecular weight | Theoretical: 559.316 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




































